 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676712750 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1033aa MW: 114049 Da PI: 8.3702 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.5 | 1.2e-40 | 132 | 209 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCevhska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+++
676712750 132 VCQVDNCTQDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTTQ 209
6**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.1E-33 | 126 | 194 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.069 | 130 | 207 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.97E-37 | 131 | 208 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.1E-28 | 133 | 206 | IPR004333 | Transcription factor, SBP-box |
| CDD | cd00204 | 1.88E-7 | 785 | 922 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-6 | 827 | 932 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.42E-7 | 828 | 930 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 8.561 | 869 | 941 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1033 aa Download sequence Send to blast |
MDEVGAQVAA PMFIHQSLSP MGRKRNLYHH QTPNRLVPSS QPQRRDEWNS KMWDWDSRRF 60 QAKPVDADVL RIGSEVQFDS TSRDKEGEER GLDLNLGCCL NVVEEETTMQ VGRPSKKVRS 120 GSPGGGGGYY PVCQVDNCTQ DLSHAKDYHR RHKVCEVHSK ATKALVGKQM QRFCQQCSRF 180 HLLSEFDEGK RSCRRRLAGH NRRRRKTTQP EEIASGVAAP GNRENTSNAN MDLMALLTAL 240 ACAQGKTEVK PVGSPAVPER EQLLQILNKI NSLPLPMDLV SKLNNIGSLA RKNLDHPVVN 300 PQNDMNGASP STMDLLAVLS ATLGSSSPDA LAILSQGGFG TKDSEKAKLS SYDQAATTNL 360 EKRTVGGERS SSSNQSPSQD SDSHAQDTRS SLSLQLFTSS PEDESRPAVA SSRKYYSSAS 420 SNPVEERSPS SSPVMQELFP LQTSPETMRS RNHQNTSPAR TGGCLPLELF GASNRGTANP 480 NFKGFGQQSG YASSGSDYSP PSLNSDSQDR TGKIVFKLLD KDPSQLPGTL RSEIYNWLSN 540 IPSEMESYIR PGCVVLSVYV AMSPAAWEQE RPVLFFFSSS KTCCNGLVLC YKILIQIFGE 600 TRGRIRCSKS WRTWNSPELI SVSPVAVVAG EETSLVVRGR SLTNDGISLR CTHMGGYESI 660 EVTGAACRRA IFDELNVNSF KVKNPQPGFL GRCFIEVENG FRGDSFPLII ANASICKELN 720 RIEEEFHPKS QDITEEQTQT SVRRPTSREE SLCFLNELGW LFQKNQTTEP RELSNFSLSR 780 FKFLLVCSVE RDYCALTRTL LGMLVERNLV NDELNREALD MLAESQLLNR AVKRKSKNMV 840 ELLVHYSVKP TALGSTKKLL FLPNITGPGG ITPLHLAACT SGSDDMVDLL TNDPQEIGLS 900 SWNSLCDAAG QTPYSYAAIR NNHTYNSLVA RKLADKRNRQ VSLNIESEIV DQLGVSRRLS 960 SEMNKSSCAT CATVALKYRR KASGSHRLFP TPIIHSMLAV ATVCVCVCVF MHAFPIVRQG 1020 SHFSWGGLDY GSI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-31 | 121 | 206 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
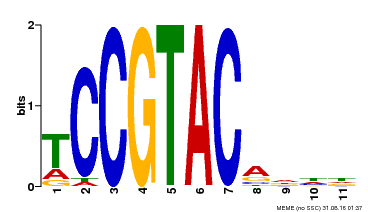
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676712750 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353554 | 0.0 | AK353554.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-52-I08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006416342.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | V4KAJ1 | 0.0 | V4KAJ1_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006416342.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



