 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676718360 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 410aa MW: 44872.2 Da PI: 7.5826 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 40 | 9.9e-13 | 195 | 241 | 1 | 51 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaark 51
s+y+GV+ +k grW+A+ + +k+++lg f+++ eAa+a+++a+
676718360 195 SKYRGVTLHK-CGRWEARMGQF--L-GKKYIYLGLFDSEVEAARAYDKAAI 241
89********.7******5553..2.26**********99*********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 10.231 | 71 | 160 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.3E-12 | 120 | 166 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.37E-6 | 121 | 160 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 8.8E-8 | 131 | 160 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 3.8E-8 | 195 | 241 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.72E-23 | 195 | 255 | No hit | No description |
| SuperFamily | SSF54171 | 3.79E-16 | 195 | 254 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.1E-15 | 196 | 254 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 15.211 | 196 | 253 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.9E-29 | 196 | 259 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-5 | 197 | 208 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.0E-5 | 235 | 255 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 410 aa Download sequence Send to blast |
MLDLNLDADS PESTQYGDSY ADRKTSDGGS GNRLEESGTS TSSVINADVA DEDSCSTRAF 60 TLNFDILKVG SSGGSSTRSW IDLSFDSHGE TKFVAPAPAP APVPAQVKKS RRGPRSRSSQ 120 YRGVTFYRRT GGFDTAHAAA RAYDRAAIKF RGVDADINFT LGDYEEDMKQ VQNLSKEEFV 180 HILRRQSTGF SRGSSKYRGV TLHKCGRWEA RMGQFLGKKY IYLGLFDSEV EAARAYDKAA 240 ISSNGREAVT NFELSSYQNE FNSDTNNEGG HDKLDLNLGI SLSPGNASKQ NGRLFQRGVN 300 LTIDNELMGK PVNTPLPYGS SDHRAYWNGA CPSYNNPVEG RATEKRSEAE GGMMSNWGWQ 360 RPGQTSAMRP QPPGAQPQLF SVAAASSGFS NFRQQPPNDN ASLGYFYPQP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). Regulates negatively the transition to flowering time and confers flowering time delay. {ECO:0000250, ECO:0000269|PubMed:14555699}. | |||||
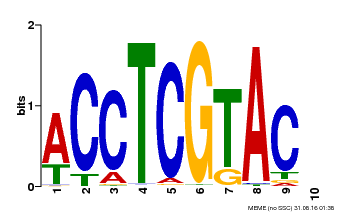
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676718360 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by miR172a-2/EAT. {ECO:0000269|PubMed:14555699}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF003100 | 1e-105 | AF003100.1 Arabidopsis thaliana AP2 domain containing protein RAP2.7 mRNA, partial cds. | |||
| GenBank | AF325074 | 1e-105 | AF325074.1 Arabidopsis thaliana putative AP2 domain transcription factor (At2g28550) mRNA, complete cds. | |||
| GenBank | AY050815 | 1e-105 | AY050815.1 Arabidopsis thaliana putative AP2 domain transcription factor (At2g28550) mRNA, complete cds. | |||
| GenBank | AY091418 | 1e-105 | AY091418.1 Arabidopsis thaliana putative AP2 domain transcription factor (At2g28550) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013631232.1 | 0.0 | PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 | ||||
| Swissprot | Q9SK03 | 0.0 | RAP27_ARATH; Ethylene-responsive transcription factor RAP2-7 | ||||
| TrEMBL | A0A3P6C1H1 | 0.0 | A0A3P6C1H1_BRAOL; Uncharacterized protein | ||||
| STRING | Bo4g070350.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5861 | 27 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 0.0 | related to AP2.7 | ||||



