 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676727272 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 387aa MW: 43384.1 Da PI: 7.2498 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 48.3 | 2.3e-15 | 88 | 132 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +W++ E+e++++a+k++G W+ I +++g +t+ q++s+ qk+
676727272 88 REKWSEKEHERFLEAIKLYGRA-WRQIQEHIG-SKTAVQIRSHAQKF 132
789*****************88.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.42E-14 | 82 | 138 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.037 | 83 | 137 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-9 | 84 | 134 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-15 | 86 | 135 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.6E-12 | 87 | 135 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-12 | 88 | 131 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.44E-9 | 90 | 133 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 387 aa Download sequence Send to blast |
MAAPVINQLP SFAKQIDELR LVYRISDKYD SLWRIICWEE VEDPSEEVSS NVENGSCNSN 60 EGTNPETSSN SLENTVKVRK PYTVTKQREK WSEKEHERFL EAIKLYGRAW RQIQEHIGSK 120 TAVQIRSHAQ KFFSKVSRET DSGSDGSTKA VAIPPPRPKR KPTHPYPRKS PLPYSQSPSS 180 AMEKGTKSPT SVLSAFASED QLNRCSSPNS CTSDMQCIDE KNDYAASKHF FKEDDAAIAS 240 TPTSSITLFG KIVVVAGESH KTSRDNVDDH KSLTGQEDHY LGITKSTSRI DSRHVDTDLS 300 LGVWETSCTG SNAFGSVTEV SENLEKSAEP INSPWKRLTS LEKQEFCYHA NGFRPYKRCL 360 SEREVTSSLS LVASEEENNR RRARVCS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phytochrome A-mediated cotyledon opening. Controlled by the central oscillator mediated by LHY and CCA1. Part of a regulatory circadian feedback loop. Regulates its own expression. {ECO:0000269|PubMed:14523250}. | |||||
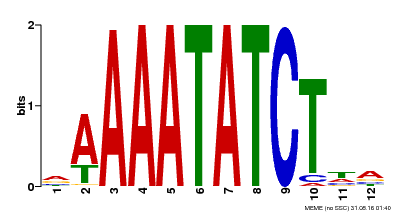
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676727272 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Peak of transcript abundance near subjective dawn. Up-regulated transiently by red light and far red light. {ECO:0000269|PubMed:14523250, ECO:0000269|PubMed:19805390}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001288892.1 | 1e-179 | protein REVEILLE 7 | ||||
| Swissprot | B3H5A8 | 1e-170 | RVE7_ARATH; Protein REVEILLE 7 | ||||
| TrEMBL | A0A3N6QIK3 | 0.0 | A0A3N6QIK3_BRACR; Uncharacterized protein | ||||
| STRING | Bra025914.1-P | 1e-179 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9174 | 27 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 1e-151 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



