 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676733544 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 346aa MW: 38773.4 Da PI: 6.1284 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 174.3 | 3.6e-54 | 20 | 146 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgelv 100
lppGfrFhPtdee++++yLk+kv + ++++ +i ++d++k ePwdLpk +k +eke yfF++rd+ky+tg r+nrat sgyWkatgkdke+++ kg lv
676733544 20 LPPGFRFHPTDEEIITHYLKEKVFNVRFTA-AAIGQADLNKNEPWDLPKIAKMGEKEFYFFCQRDRKYPTGMRTNRATLSGYWKATGKDKEIFRGKGCLV 118
79*************************999.78***************8888899***************************************9***** PP
NAM 101 glkktLvfykgrapkgektdWvmheyrl 128
g+kktLvfy+grapkgekt+Wvmheyrl
676733544 119 GMKKTLVFYRGRAPKGEKTNWVMHEYRL 146
**************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.18E-62 | 17 | 172 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 58.234 | 20 | 171 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.7E-28 | 21 | 146 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:1900057 | Biological Process | positive regulation of leaf senescence | ||||
| GO:1903648 | Biological Process | positive regulation of chlorophyll catabolic process | ||||
| GO:0000975 | Molecular Function | regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 346 aa Download sequence Send to blast |
MVEEVCVVVN QGGDQEAVDL PPGFRFHPTD EEIITHYLKE KVFNVRFTAA AIGQADLNKN 60 EPWDLPKIAK MGEKEFYFFC QRDRKYPTGM RTNRATLSGY WKATGKDKEI FRGKGCLVGM 120 KKTLVFYRGR APKGEKTNWV MHEYRLDGKY SYHNLPKSAR DEWVVCRVFH KNAPPPTTTT 180 ITTHTTNQLI RIESFDNIDH LLDFSSLPPL IDSGFLGQPG PSFSGASQQD DRKPILHHPT 240 TAPIDNTYIP AQTLSYPYNH SVQNSASCLG YGTGSGNNNN AMIKLEHSLV SVSQETGLSS 300 DVNTTATPEI SSYPMMMNSA ANAAMMDGNN TSYDDDDDLG IFWDDY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 4e-50 | 18 | 173 | 15 | 167 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 4e-50 | 18 | 173 | 15 | 167 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 4e-50 | 18 | 173 | 15 | 167 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 4e-50 | 18 | 173 | 15 | 167 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swm_B | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swm_C | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swm_D | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swp_A | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swp_B | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swp_C | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 3swp_D | 5e-50 | 18 | 173 | 18 | 170 | NAC domain-containing protein 19 |
| 4dul_A | 4e-50 | 18 | 173 | 15 | 167 | NAC domain-containing protein 19 |
| 4dul_B | 4e-50 | 18 | 173 | 15 | 167 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that acts as positive regulator of leaf senescence. Activates NYC1, SGR1, SGR2 and PAO, which are genes involved in chlorophyll catabolic processes. Activates senescence-associated genes, such as RNS1, SAG12 and SAG13. {ECO:0000269|PubMed:27021284}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00332 | DAP | Transfer from AT3G04060 | Download |
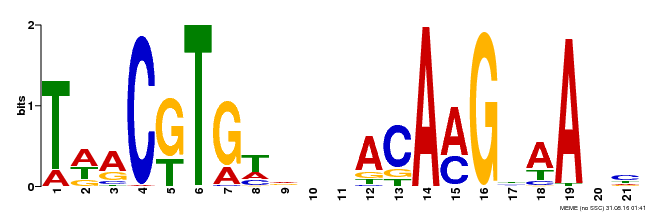
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676733544 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF361623 | 0.0 | AF361623.1 Arabidopsis thaliana AT3g04060/T11I18_17 mRNA, complete cds. | |||
| GenBank | AY081834 | 0.0 | AY081834.1 Arabidopsis thaliana AT3g04060/T11I18_17 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_187056.1 | 0.0 | NAC domain containing protein 46 | ||||
| Swissprot | Q9SQQ6 | 0.0 | NAC46_ARATH; NAC domain-containing protein 46 | ||||
| TrEMBL | A0A1J3CDP6 | 0.0 | A0A1J3CDP6_NOCCA; Uncharacterized protein | ||||
| STRING | AT3G04060.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2269 | 27 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04060.1 | 0.0 | NAC domain containing protein 46 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



