 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676734844 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 939aa MW: 104976 Da PI: 5.9436 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 135.2 | 2.1e-42 | 148 | 225 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C+adls++k+yhrrhkvCe+hska++++v g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
676734844 148 VCQVENCDADLSKVKDYHRRHKVCEMHSKATSAIVGGIMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTNP 225
6**************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.7E-34 | 143 | 210 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.599 | 146 | 223 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.49E-39 | 148 | 227 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.7E-31 | 149 | 222 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 8.86E-8 | 729 | 830 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.7E-8 | 731 | 828 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.91E-9 | 731 | 827 | No hit | No description |
| PROSITE profile | PS50297 | 8.694 | 731 | 827 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 939 aa Download sequence Send to blast |
MEARVEDEGQ HFYGFRAMDL RSVGKKSVEW DLNDWKWDGD LFVATQLNPG ASETIGRQFF 60 PIGNSSNSSS SCSDEGNNHN NTESTVRGTR EIERKRRAAT TGEENNNGGG GGLTLKLGDN 120 GYDLNGDREG NSAAKKTKFG GGVTNRSVCQ VENCDADLSK VKDYHRRHKV CEMHSKATSA 180 IVGGIMQRFC QQCSRFHVLQ EFDEGKRSCR RRLAGHNKRR RKTNPEPGAN GNPLSDDNQS 240 SNYLLISLLK ILSNMHSNRS DHPGDQDLMS HLLKSLVSHA GEQLGKNLVE LLLQGGSQAS 300 LNIGNSGLLA IDQAPQEDLK QTPEIPRQEL YANGTATENR SEKRAKVNDF DLNDIYIDSD 360 DGTYIERSPP PTNPATSSLD YPSWIHQSSP PQTSRNSDSA SDQSPSSSSE DAQMRTGRIV 420 FKLFGKEPND FPVVLRGQIL DWLSHSPTDM ESYIRPGCIV LTIYLRQAET AWEELSDDMG 480 FSLSKLLALS DDPLWTTGWI YVRVQNQLAF VFDGQVVVDT SLPLRSRDYS HIINVRPLAV 540 AATGKAQFTV KGINLRRPGT RLLCAVEGKY LIEETAHDSA KENDDLKENS EIVECVNFSC 600 DLPITSGRGF MEIEDQGLSS SFFPFIVLEE DDVCSEIRIL ETTLEFTETD SAKQAMEFIH 660 EMGWLLHRSK LGESDPNPDV FPLTRFKWLI EFSMDREWCA VIRKLLNMFF EGAVGDFSSS 720 SDAELSELCL LHRAVRKNSK PMVQMLLRYV PKQQRQSLFR PDAAGPAGLT PLHIAAGKDG 780 SEDVLDALTE DPAMVGIEAW RTSRDSTGFT PEDYARLRGH FSYIHLIQRK INKKSTTEDH 840 VVVNIPVSFS DREQKERKSG PMASALEITH ANHQLQCKLC DHKLVYGTAR RSVAYRPAML 900 SMVAIAAVCV CVALLFKSCP EVLYVFQPFR WELLDYGTR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-32 | 142 | 222 | 4 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
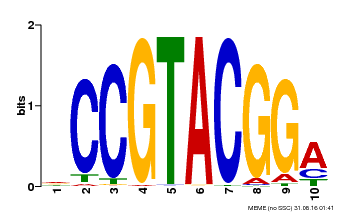
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676734844 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC815948 | 0.0 | KC815948.1 Arabis alpina SQUAMOSA promoter binding protein-like protein 1 (SPL1) gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006397898.1 | 0.0 | squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | V4N2D6 | 0.0 | V4N2D6_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006397898.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



