 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676735874 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 313aa MW: 35180.2 Da PI: 5.986 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 163.8 | 6.1e-51 | 14 | 139 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgelv 100
lppGfrF Ptdeel+v+yL++k++g +++l + i+e+d+yk++Pw Lp+k+ +ekewyfFs+rd+ky++g+r+nr++ sgyWkatg+dk + + +g+ v
676735874 14 LPPGFRFYPTDEELMVQYLCRKAAGYDFSL-QLIAEIDLYKFDPWVLPNKALFGEKEWYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKIIST-EGKRV 111
79***************************9.89***************8888899*********************************999988.9999* PP
NAM 101 glkktLvfykgrapkgektdWvmheyrl 128
g+kk Lvfy g+apkg+kt+W+mheyrl
676735874 112 GIKKALVFYIGKAPKGTKTNWIMHEYRL 139
**************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.05E-66 | 9 | 162 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.675 | 14 | 162 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.4E-25 | 15 | 139 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 313 aa Download sequence Send to blast |
MGIQETDPLS QLSLPPGFRF YPTDEELMVQ YLCRKAAGYD FSLQLIAEID LYKFDPWVLP 60 NKALFGEKEW YFFSPRDRKY PNGSRPNRVA GSGYWKATGT DKIISTEGKR VGIKKALVFY 120 IGKAPKGTKT NWIMHEYRLI EPSRTNGSSK LDDWVLCRIY KKQASAQKQA YENVNKSTTE 180 LSNNGTSSTT SSSSHFEDVL DSLHHEIDNR NFQFPNSNRF SSVRPELTAE KTGFNGLADG 240 SSFDWGSFIG NVEHNSGPEL GMSHVVPNLE FNSGYLKMEE EFINNSDELV QNGYGVDSVG 300 FGYPGQVGGF GFI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-121 | 1 | 168 | 4 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-121 | 1 | 168 | 4 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-121 | 1 | 168 | 4 | 171 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-121 | 1 | 168 | 4 | 171 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 1e-121 | 1 | 168 | 7 | 174 | NAC domain-containing protein 19 |
| 4dul_A | 1e-121 | 1 | 168 | 4 | 171 | NAC domain-containing protein 19 |
| 4dul_B | 1e-121 | 1 | 168 | 4 | 171 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factors that bind specifically to the 5'-CATGTG-3' motif. {ECO:0000269|PubMed:15319476}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00012 | PBM | Transfer from AT1G52890 | Download |
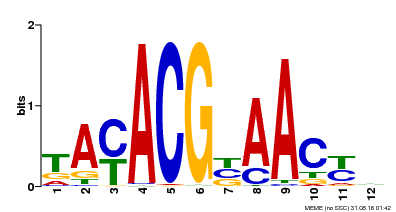
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676735874 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by drought, high salinity and abscisic acid (ABA). Slightly up-regulated by jasmonic acid. Not induced by cold treatment. {ECO:0000269|PubMed:12646039, ECO:0000269|PubMed:15319476}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KC966747 | 0.0 | KC966747.1 Brassica napus NAC transcription factor 19 (NAC19.1) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009147649.1 | 0.0 | PREDICTED: NAC domain-containing protein 19 | ||||
| Swissprot | Q9C932 | 0.0 | NAC19_ARATH; NAC domain-containing protein 19 | ||||
| TrEMBL | A0A219UXT7 | 0.0 | A0A219UXT7_BRAJU; NAC domain protein 19 | ||||
| STRING | XP_006392855.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1435 | 27 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G52890.1 | 0.0 | NAC domain containing protein 19 | ||||



