 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676739168 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 920aa MW: 104730 Da PI: 7.2648 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 162.6 | 7.3e-51 | 30 | 146 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywl 101
+e +rwl+++ei+a+L n++ +++ ++ + pksg+++L++rk++r+frkDG++wkkkkdgkt++E+he+LKvg+ e +++yYah+++nptf rrcywl
676739168 30 EEaYSRWLRPNEIHALLCNHKYFTINVKPVNLPKSGTIVLFDRKMLRNFRKDGHNWKKKKDGKTIKEAHEHLKVGNEERIHVYYAHGDDNPTFVRRCYWL 129
55589*********************************************************************************************** PP
CG-1 102 Leeelekivlvhylevk 118
L++++e+ivlvhy+e++
676739168 130 LDKSKEHIVLVHYRETH 146
**************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 76.347 | 25 | 151 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.3E-74 | 28 | 146 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.1E-45 | 32 | 144 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.49E-13 | 372 | 458 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 2.17E-13 | 558 | 668 | No hit | No description |
| Pfam | PF12796 | 9.7E-7 | 559 | 638 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-15 | 559 | 671 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 3.26E-15 | 567 | 674 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 13.761 | 576 | 641 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.808 | 609 | 641 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2.7E-6 | 609 | 638 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50096 | 7.181 | 758 | 784 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 330 | 773 | 795 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.9E-4 | 796 | 818 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.347 | 797 | 821 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 11 | 872 | 894 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.279 | 873 | 902 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 920 aa Download sequence Send to blast |
MAGVDSGRLI GSEIHGFHTL QDLDIQTMME EAYSRWLRPN EIHALLCNHK YFTINVKPVN 60 LPKSGTIVLF DRKMLRNFRK DGHNWKKKKD GKTIKEAHEH LKVGNEERIH VYYAHGDDNP 120 TFVRRCYWLL DKSKEHIVLV HYRETHEVQA APATPGNSYS SSITDHLSPI PVAEDINSGV 180 RNACNTGFEA RDNSLVAKSH EIRLHEINTL DWDELLVSAD SQCLPTEEDV LYFTDQLETA 240 PRGSAKPGNH LVGYNASTDL PSFLGLGDPV YQNNNPCGAR EFSSQHLHCV VDPNLQRRDS 300 TEMVADEPGD ALLNNGYGSQ ESFGKWVNNF ISDSPGSVED PSLEAVFTPG QESSAPPAVL 360 HSQSNIPEQV FNITDVSPAW AYSTEKTKIL VTGFFHDSFQ HFGRSNLFCI CGELRVPAEF 420 LQMGVYRCFL PPQSPGVVNL YLSADGNKPI SQLFSFEHRS VPVIEKVVPQ EDQLYKWEEF 480 EFQVRLAHLL FTSSNKINVF ASKISPDNLL EAKKLASRTS HLLNSWAYLM KSIQANEVPF 540 DQARDHLFEL TLKNRLKEWL LEKVIENRNT KEYDSKGLGV IHLCAVLGYT WSILLFSWAN 600 ISLDFRDKHG WTALHWAAYY GREKMVAALL SAGARPNLVT DPTKEFLGGC TAADLAQQKG 660 YEGLAAFLAE KCLIAQFEDM RLAGNISGNL EAIKAETSTN PGHSNEEEQS LKDTLAAYRT 720 AAEAAARIQG AFREHELKVR SNAVRFASKE EEAKNIIAAM KIQHAFRNYE TRRKIAAAAR 780 IQYRFQTWKI RREFLNMRKK AIRIQAAFRG YQVRRQYQKI TWSVGVLEKA ILRWRLKRKG 840 FRGLQLSQPE EKEGTEVVED FFKTSQKQAE DRLERSVVRV QAMFRSKKAQ QDYRRMKLAH 900 EQAQLEYDGM QEFDEMAMES |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
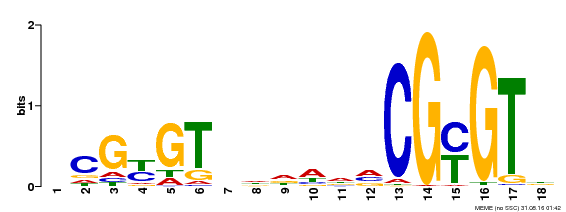
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676739168 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY128295 | 0.0 | AY128295.1 Arabidopsis thaliana AT4g16150/dl4115w mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013711273.1 | 0.0 | calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A3P6GMJ4 | 0.0 | A0A3P6GMJ4_BRAOL; Uncharacterized protein | ||||
| STRING | Bo8g043280.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



