 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676739534 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1071aa MW: 119488 Da PI: 6.49 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 127 | 7.6e-40 | 65 | 137 | 36 | 108 |
CG-1 36 sgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywlLeeelek 108
gsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+e+n++fqrrcyw+Le+ +
676739534 65 GGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEDNENFQRRCYWMLEHFSAD 137
69*****************************************************************975332 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 57.679 | 37 | 164 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.0E-49 | 40 | 159 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.1E-31 | 65 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 7.4E-15 | 488 | 574 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 5.1E-4 | 488 | 562 | IPR002909 | IPT domain |
| PROSITE profile | PS50297 | 10.365 | 671 | 720 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.45E-8 | 672 | 771 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-8 | 672 | 772 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.20E-6 | 677 | 769 | No hit | No description |
| PROSITE profile | PS50088 | 9.351 | 688 | 720 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.64 | 884 | 906 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 1.19E-7 | 885 | 934 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.95 | 885 | 914 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0021 | 886 | 905 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 907 | 929 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.413 | 908 | 932 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.7E-4 | 910 | 929 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1071 aa Download sequence Send to blast |
MIQQKDTHMH KIPSVYPRIS SSCCPKHNTG GLGLLRFVRF FGTIRSFILR QSLPTDHPTF 60 RALLGGSLFL FDRKVLRYFR KDGHNWRKKK DGKTVKEAHE KLKVGSIDVL HCYYAHGEDN 120 ENFQRRCYWM LEHFSADVLC FYGYRDLMHI VFVHYLEVKG NRVSSSGIRE KNSNSLSGSA 180 SVNIDSTATP SSALSPLCED ADSGDSRQAS SSLQANREPQ TVASQIRHQQ NANTINSYNP 240 TSTLGNRGGW PSAPGIGFVS QVHGNRVKES DSQRSVGVPA WDASFENPLA RYQNLPYNAL 300 LTQTQPPSAG LIPVEGNTEK GSLLTAEHRR NPLQNEVNWQ VPVQDSLPSQ KWSMDSHSAM 360 ADDTDLALHE NFGTFSSLLG NQNQQLIGGS FQAPFTSTEA AYIPKLGPED LLYEASANQS 420 LPLRKSLLKK EDSLKKVDSF SRWVSNELSE MEDLHMQSSS GGLAWTSVET AAAASSLSPS 480 LSEDQRFTII DFWPKWSQTD SEVEVMVIGT FLLSPQEVSS YSWSCMFGEV EVPAEILVDG 540 VLCCHAPPHE VGQVPFYITC SDRFSCSEVR EFDFLPGSAK KLNTADIYGA FTNEASLHLR 600 FENLLARISS VQEQHIFEDV GEKRRKISRI MLLKEEKDTL FPGTVVKDPT ELEAKERLIR 660 EEFEDKLYLW LIHKVTEEGK GPNILDEEGQ GVLHLAAALG YDWAIKPILA AGVSINFRDA 720 NGWEDTVALL VSLGADSGAL TDPSPELPLG KAASDLAYGN GHRGISGFLA ESSLTSYLEK 780 LTVDAKENSS ADSSRAKAVQ TVAERTATPM SDGDVPETLS MKDSITAVFN ATQAADRLHQ 840 VFRMQSFQRK QLSEFGDNEF DISDELAVSF AAAKTKKPGH SSGAAHAAAV QIQKKYRGWK 900 KRKEFLLIRQ RIVKIQAHVR GHQVRKQYRT IIWSVGLLEK IILRWRRKGS GLRGFKRDTI 960 TKAPEPVCAP PQEDDYDFLK EGRKQTEERL QKALTRVKSM AQYPEARAQY RRLLTVVEGI 1020 RENEASSSSA MNNNNNNNSN TEEAANYNKE DDLIDIDSLL DDDTFMSLAF E |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
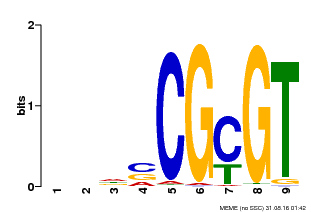
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676739534 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB493811 | 0.0 | AB493811.1 Arabidopsis thaliana At5g64220 mRNA for hypothetical protein, partial cds, clone: RAAt5g64220. | |||
| GenBank | AK229403 | 0.0 | AK229403.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 2, complete cds, clone: RAFL16-66-C08. | |||
| GenBank | BT010874 | 0.0 | BT010874.1 Arabidopsis thaliana At5g64220 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006394184.1 | 0.0 | calmodulin-binding transcription activator 2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A1J3JZU7 | 0.0 | A0A1J3JZU7_NOCCA; Calmodulin-binding transcription activator 2 | ||||
| STRING | XP_006394184.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



