 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676742326 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 373aa MW: 40544.6 Da PI: 8.1749 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 137.1 | 5.4e-43 | 72 | 149 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+CqvegC +dl++ak y++rh+vC +hsk+p+v+v+g+eqrfCqqCsrfh+l efD ekrsCrrrLa+hnerrrk+q+
676742326 72 RCQVEGCGMDLTNAKGYYSRHRVCGMHSKTPKVVVAGIEQRFCQQCSRFHQLPEFDLEKRSCRRRLAGHNERRRKPQP 149
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.8E-34 | 65 | 134 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 31.923 | 70 | 147 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.01E-40 | 71 | 151 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 2.2E-32 | 73 | 146 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:2000025 | Biological Process | regulation of leaf formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 373 aa Download sequence Send to blast |
MDMGSNSGPG LGPGQAESGG SSTESSSFSG GLMFGQKIYF EDAGGGSGSS PSGGSNRRGR 60 GGASGHSGQI PRCQVEGCGM DLTNAKGYYS RHRVCGMHSK TPKVVVAGIE QRFCQQCSRF 120 HQLPEFDLEK RSCRRRLAGH NERRRKPQPA SLSVLSSRYG RIAPSLYGNA ETTMSGSFLG 180 NQEMGWTSAR TLDTRVMRRP VSAPSWQINP MNVFSQGSVG GGGISFSSPD ILDTKPESYK 240 GIGDSNCALS LLSNPHQPQD NNNNNNNAWR TSSGFGPMTV TMAQPPPAPN QHQYLHPPWV 300 FKDDDNNCPS DMSPVLNLGR FTETDNCQMS GGTTMGEFEL SDNHHHHQNR RQYMEADNTR 360 AYDSSSHHNN WSL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-29 | 62 | 146 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
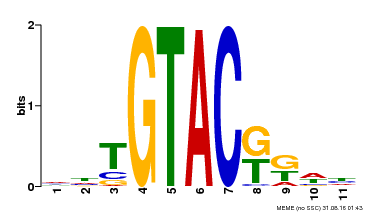
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00307 | DAP | Transfer from AT2G42200 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676742326 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156 and miR157. {ECO:0000305|PubMed:12202040}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353538 | 0.0 | AK353538.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-50-J24. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006411480.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q700W2 | 0.0 | SPL9_ARATH; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | E4MYE1 | 0.0 | E4MYE1_EUTHA; mRNA, clone: RTFL01-50-J24 | ||||
| TrEMBL | V4MJX0 | 0.0 | V4MJX0_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006411480.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2144 | 27 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42200.1 | 0.0 | squamosa promoter binding protein-like 9 | ||||



