 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676750136 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | GATA | ||||||||
| Protein Properties | Length: 304aa MW: 33513 Da PI: 4.9607 | ||||||||
| Description | GATA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | GATA | 44.4 | 2.3e-14 | 200 | 227 | 8 | 35 |
GATA 8 kTplWRrgpdgnktLCnaCGlyyrkkgl 35
Tp+WR gp g+ktLCnaCG++y++ +l
676750136 200 ATPQWRTGPMGPKTLCNACGVRYKSGRL 227
4***********************9885 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF016992 | 6.2E-68 | 1 | 281 | IPR016679 | Transcription factor, GATA, plant |
| SMART | SM00401 | 4.1E-6 | 193 | 237 | IPR000679 | Zinc finger, GATA-type |
| Gene3D | G3DSA:3.30.50.10 | 2.8E-10 | 199 | 225 | IPR013088 | Zinc finger, NHR/GATA-type |
| Pfam | PF00320 | 4.3E-12 | 200 | 227 | IPR000679 | Zinc finger, GATA-type |
| SuperFamily | SSF57716 | 2.71E-10 | 200 | 251 | No hit | No description |
| CDD | cd00202 | 1.75E-10 | 201 | 239 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 304 aa Download sequence Send to blast |
MEEEAHEFFL TADHFAVDDL LVDFSNDDDD ENDVIVDSDG TNTALAVADS SNSSSFSALG 60 LPNFHSDVQD GTSFSGDLCV PSDELAELEW LSNFVEDSFS TEDVQKLQLI SGYKARPEPK 120 SEPGPENPNS SSPIFATDVS VPAKARSKRS RAAACNWASR GLLMETVYDS PFTGETILSG 180 HNLSPPTSPT SMTQLGKKQA TPQWRTGPMG PKTLCNACGV RYKSGRLVPE YRPAASPTFV 240 MTKHSNSHRK VMELRRQKEM TKAHHEFIHH HHHGTDTAMM FDVSSDGDDY LIHHNVGPDF 300 RQLI |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that specifically binds 5'-GATA-3' or 5'-GAT-3' motifs within gene promoters. May be involved in the regulation of some light-responsive genes (By similarity). Transcription activator involved in xylem formation. Functions upstream of NAC030/VND7, a master switch of xylem vessel differentiation (PubMed:25265867). {ECO:0000250|UniProtKB:Q8LAU9, ECO:0000269|PubMed:25265867}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00530 | DAP | Transfer from AT5G25830 | Download |
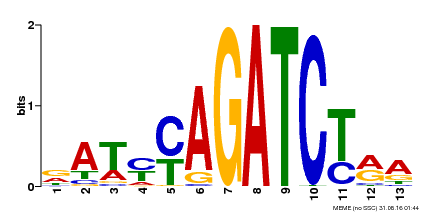
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676750136 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC005405 | 1e-106 | AC005405.1 BAC F18A17 from chromosome V containing TINY at 60.5 cM, complete sequence. | |||
| GenBank | CP002688 | 1e-106 | CP002688.1 Arabidopsis thaliana chromosome 5 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009151105.1 | 0.0 | PREDICTED: GATA transcription factor 12 | ||||
| Refseq | XP_013643773.1 | 0.0 | GATA transcription factor 12 | ||||
| Swissprot | P69781 | 1e-155 | GAT12_ARATH; GATA transcription factor 12 | ||||
| TrEMBL | A0A1J3FP63 | 0.0 | A0A1J3FP63_NOCCA; GATA transcription factor | ||||
| STRING | Bra009864.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6625 | 26 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G25830.1 | 1e-116 | GATA transcription factor 12 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



