 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676751498 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 588aa MW: 66364.7 Da PI: 5.0712 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 439.3 | 3.4e-134 | 40 | 403 | 1 | 351 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkekve 100
e+l++rmwkd+++lkr+ker+k+ + a ++ +k+++qa+rkkmsraQDgiLkYMlk mevc+++GfvYgiipekgkpv+g+sd++raWWkekv+
676751498 40 EDLERRMWKDRVRLKRIKERQKSGS--QG-AQTKEMPKKISDQAQRKKMSRAQDGILKYMLKLMEVCKVRGFVYGIIPEKGKPVSGSSDNIRAWWKEKVK 136
89********************965..33.35567789************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----GGG CS
EIN3 101 fdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkphd 200
fd+ngpaai+ky+ ++l++++++++ ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekg++pPWWPtG+e+ww +lgl+k+q+ ppy+kphd
676751498 137 FDKNGPAAIAKYEEECLAFGKSDGN----RNSQFVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGTPPPWWPTGNEEWWVKLGLPKSQS-PPYRKPHD 231
*******************987777....77999*********************************************************.9******* PP
--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX...XX.....XXXX....XXXXXXXXXXXXXXXXXXX CS
EIN3 201 lkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah...ss.....slrk....qspkvtlsceqkedvegkk 288
lkk+wkv+vLtavi+hmsp+i++i++++rqsk+lqdkm+akes+++l+vlnqee+++++ s++ s+ + +++k+++++++++dv+g++
676751498 232 LKKMWKVGVLTAVINHMSPDIAKIKRHVRQSKCLQDKMTAKESAIWLAVLNQEESLIQQPSSDngtSNvtethR--RgnnaDRRKTVINSDSDYDVDGTE 329
***************************************************************65422787540..33445888999**********999 PP
XXX.XXXXXXXXXX..................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXXXXX CS
EIN3 289 eskikhvqavktta..................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgsetelifadknsisq 351
e + + ++++++++ +++ +r+rk+++s+s +v + q+++ ++ e+++i++d+n+++
676751498 330 EASGSVSSKDNRRNqiqkeqptatsqpvsdqdKVEKHRRRKRPRSRSGTV-------NRQEEEQPEAEQRNILPDMNHVDA 403
99999999999888999999999966555444444444444333333333.......345566667899999999999875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.9E-126 | 40 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.7E-71 | 161 | 293 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 6.8E-61 | 166 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 588 aa Download sequence Send to blast |
MGGGDPAPSG ADIRMENEPD DLASDNVAEI DVSDEEIDAE DLERRMWKDR VRLKRIKERQ 60 KSGSQGAQTK EMPKKISDQA QRKKMSRAQD GILKYMLKLM EVCKVRGFVY GIIPEKGKPV 120 SGSSDNIRAW WKEKVKFDKN GPAAIAKYEE ECLAFGKSDG NRNSQFVLQD LQDATLGSLL 180 SSLMQHCDPP QRKYPLEKGT PPPWWPTGNE EWWVKLGLPK SQSPPYRKPH DLKKMWKVGV 240 LTAVINHMSP DIAKIKRHVR QSKCLQDKMT AKESAIWLAV LNQEESLIQQ PSSDNGTSNV 300 TETHRRGNNA DRRKTVINSD SDYDVDGTEE ASGSVSSKDN RRNQIQKEQP TATSQPVSDQ 360 DKVEKHRRRK RPRSRSGTVN RQEEEQPEAE QRNILPDMNH VDAPLLDYNI NDTHHHEEGV 420 LEPNIALGPE ENGLELVVPE FDNNYTYLPP VNGQAMMPVD ERPMLYGSNP NQELQFGSGY 480 NYYNPSAVFV HNQEEDIIHT PIEINAQAQP HNNGFEAPGG VLRPHDLLGN EDGVAGRDLP 540 PQFPSDQEKL LDSNILSPFN DLTFDSSTFY SGFDSYGAFD DDYSWFGA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 5e-86 | 165 | 296 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
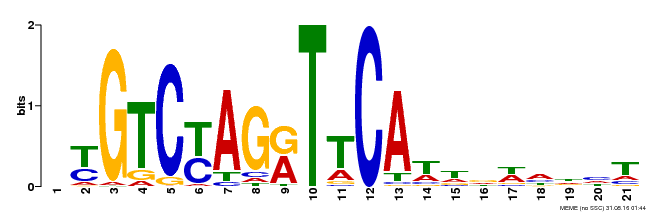
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676751498 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK353245 | 0.0 | AK353245.1 Thellungiella halophila mRNA, complete cds, clone: RTFL01-23-B16. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006390517.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | E4MXJ8 | 0.0 | E4MXJ8_EUTHA; mRNA, clone: RTFL01-23-B16 | ||||
| TrEMBL | V4KKK7 | 0.0 | V4KKK7_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006390517.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4658 | 25 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 0.0 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



