 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676754416 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 318aa MW: 35695 Da PI: 8.4147 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 103.6 | 1.2e-32 | 84 | 139 | 3 | 59 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRrevee 59
+++YkeClkNhAas+Gg+a+DGCgEfmps ge+g+++al+C+AC+CHRnFHRre e
676754416 84 TIKYKECLKNHAASMGGNAIDGCGEFMPS-GEDGSIEALTCSACNCHRNFHRREIEG 139
689*************************9.999*********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 2.0E-26 | 85 | 147 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 1.4E-29 | 85 | 137 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 6.7E-32 | 86 | 137 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 27.822 | 87 | 136 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| SuperFamily | SSF46689 | 5.83E-19 | 215 | 285 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.4E-30 | 221 | 288 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 5.4E-26 | 226 | 282 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 318 aa Download sequence Send to blast |
MEVASQADHD MPIPMNTTYG NSYGAHGHVI HHHHHPANST APNPLIVPSN GNGLDKNHDH 60 PPPHRHHVGY NIMLSNNKNE KPVTIKYKEC LKNHAASMGG NAIDGCGEFM PSGEDGSIEA 120 LTCSACNCHR NFHRREIEGE EKTFFSPYHH NQPPQPQRKL MFHHHHHKMV RSPLPQQMVM 180 PIGVATAAGS NSESEDHMEE DGGGNLTFRQ PPPPPAPYSY GGHNQKKRFR TKFTQAQKEK 240 MLSFAERIGW KMQRQEESVV QQFCQEIGVR RRVLKVWMHN NKHILSKKSS NVNNDVELSA 300 GNNDTNNKAP LGDLVSSP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh7_A | 2e-23 | 227 | 283 | 18 | 74 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. {ECO:0000250}. | |||||
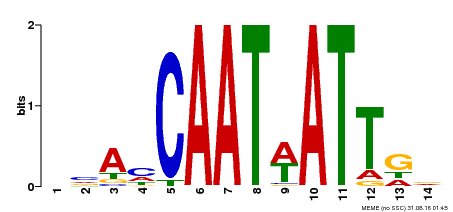
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00258 | DAP | Transfer from AT2G02540 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676754416 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018483487.1 | 1e-176 | PREDICTED: zinc-finger homeodomain protein 3-like | ||||
| Refseq | XP_018483488.1 | 1e-176 | PREDICTED: zinc-finger homeodomain protein 3-like | ||||
| Swissprot | O64722 | 1e-149 | ZHD3_ARATH; Zinc-finger homeodomain protein 3 | ||||
| TrEMBL | V4KST6 | 1e-172 | V4KST6_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006395776.1 | 1e-173 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3339 | 26 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02540.1 | 1e-152 | homeobox protein 21 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



