 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676776274 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | M-type_MADS | ||||||||
| Protein Properties | Length: 303aa MW: 34133.6 Da PI: 4.9606 | ||||||||
| Description | M-type_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 40.7 | 3.1e-13 | 12 | 48 | 2 | 37 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-.EEEE CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCda.evav 37
+ienks ++vtf+kRr g++ KA EL+ L a ++a+
676776274 12 KIENKSVKSVTFTKRRDGLFRKAAELCLLSSAsQIAI 48
69***************************87624555 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50066 | 15.699 | 3 | 51 | IPR002100 | Transcription factor, MADS-box |
| SMART | SM00432 | 1.1E-12 | 3 | 64 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00266 | 7.53E-22 | 4 | 80 | No hit | No description |
| SuperFamily | SSF55455 | 2.35E-15 | 5 | 78 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.8E-9 | 5 | 25 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 6.9E-12 | 12 | 53 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 2.8E-9 | 25 | 40 | IPR002100 | Transcription factor, MADS-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 303 aa Download sequence Send to blast |
MGGTKRKIEI VKIENKSVKS VTFTKRRDGL FRKAAELCLL SSASQIAILA TPPSSNSHAS 60 FYSFGHSSVD HVVSSLLNGK SPLPTNQVNK SGLGFWWENE GFDRSENIED LKEATDAISR 120 MLNKLRVQLD AMQSNQRDGG LVMHQDEVIQ LGNTSTNNNE EMMNQITKFE GTSGRRNPEM 180 VRLKKLPIDP QCREVGAVED EVTFKEEIVS DDGSRDQREV VPDDDCEAIG DDPADEDELE 240 ENVVKKKWSK TYQPNFKILR TMIEWTAMIV GTMSYGMRIL YLIEREKRRE SLPVCVILAS 300 IGD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00208 | DAP | Transfer from AT1G60920 | Download |
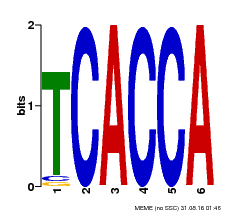
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676776274 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018454191.1 | 1e-90 | PREDICTED: agamous-like MADS-box protein AGL97 | ||||
| Refseq | XP_018454192.1 | 1e-90 | PREDICTED: agamous-like MADS-box protein AGL97 | ||||
| TrEMBL | A0A0D3E567 | 1e-87 | A0A0D3E567_BRAOL; Uncharacterized protein | ||||
| STRING | Bo9g043530.1 | 2e-88 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM454 | 17 | 145 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G60920.1 | 3e-69 | AGAMOUS-like 55 | ||||



