 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676776926 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1422aa MW: 159589 Da PI: 7.2708 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 13 | 0.0003 | 1330 | 1352 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp Cgk F ++ +L++H r+H
676776926 1330 CPvkGCGKNFFSHKYLVQHRRVH 1352
9999*****************99 PP
| |||||||
| 2 | zf-C2H2 | 10.7 | 0.0017 | 1388 | 1414 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C Cg++F+ s++ rH r+ H
676776926 1388 YVCAepRCGQTFRFVSDFSRHKRKtgH 1414
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 2.5E-16 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 13.952 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 8.1E-14 | 21 | 54 | IPR003349 | JmjN domain |
| SMART | SM00558 | 7.7E-52 | 199 | 368 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 33.593 | 202 | 368 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 8.51E-27 | 214 | 386 | No hit | No description |
| Pfam | PF02373 | 8.1E-38 | 232 | 351 | IPR003347 | JmjC domain |
| SMART | SM00355 | 12 | 1305 | 1327 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.016 | 1328 | 1352 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.965 | 1328 | 1357 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 3.5E-6 | 1329 | 1356 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1330 | 1352 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.35E-10 | 1344 | 1386 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.0E-8 | 1357 | 1381 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0015 | 1358 | 1382 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.928 | 1358 | 1387 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1360 | 1382 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 6.18E-8 | 1375 | 1410 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.0E-8 | 1382 | 1411 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 9.5 | 1388 | 1414 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.281 | 1388 | 1419 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1390 | 1414 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1422 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPVA PEFRPTLAEF QDPIAYIFKI EEEASRYGIC KILPPVPPSS 60 KKTAINNLNR SLAARAAARV REGGRASDYD DGPTFATRQQ QIGFCPRKQR PVQRPVWQSG 120 EHYTFGEFEF KAKSFEKNYL KKCGKKSSVS ALEMETLYWR ATVDKPFSVE YANDMPGSAF 180 IPLSLAAARR RESGGDGGTV GETGWNMRAM ARAEGSLLQF MKEEIPGVTS PMVYIAMMFS 240 WFAWHVEDHD LHSLNYLHMG ASKTWYGVPK DAAVAFEEVV RVHGYGEELN PLVTFSTLGE 300 KTTVMSPEVF VKAGIPCCRL VQNPGDFVVT FPGAYHSGFS HGFNFGEASN IATPQWLKIA 360 KDAAIRRAAI NYPPMVSHLQ LLYDYALALG SRVPVSIHTK PRSSRLKDKK RSEGEKLTKE 420 LFVQNIIHNN ELLHSLGKGS SIALLPQSSS DISVCSDLRI GSHLSDNQEK TIQLKSEDLS 480 SDSVTVGVSN VVKDAVSVKE KFTTLCERNR NHLVSKENET RGTSADSERR KSDRAVGLSD 540 QRLFSCVTCG VLSFDCVAIV QPKEAAARYL MSGDCSFFND WTVPSGSATL GQVAGSHHQQ 600 GTENPDVNNF YNVPGQTADH SMKTGDQITL SSSLTKANKD NGALGLLASA YGDSSDSEED 660 DHKGLDIPVS EEDACVLEAS SFDTDGKEET RDGLSSSFNS QRLTCEKGKE ADVSHATSSC 720 STLSCTSEQN RLSKGGNTSP LEITLPFISR SDDDSSRLHV FCLEHAAEVE QRLRPIGGIN 780 IMLLCHPEYP RIEAEGKIVA EELVLDHEWS DTEFRNVTRE DEETIQAALA NVEAKAGNSD 840 WAVKLGINLS YSAILSRSPL YSKQMPYNSV VYNAFGRSSL ATSSPTKSQV SGKRSSRQRK 900 YVAGKWCGKV WMSHQVHPFL LEEDLEEEES ERSHLRDEDV NGKRLFPCND SRDATTMFGR 960 KYCRKRKRRA KTVPRKKLSS FKREEGLSDD TSEDHSYKQQ WRASGNEEEL SDDSSNQMSD 1020 QQQLKEIVIH QEYHSDDEVS DRSLGQEYAV RECAASESSM ENGFQLYREK HPMYGDDDDD 1080 DDIYRHPRGI PRSKRTKVFR NRVSYDSEEN GVYQQRGRVY TSNAQTNRVV GEYDSVENSL 1140 EEHDFCSTGK RQTRSTAKRK VKTEIIQNLK DTRGRTLQQS GSRKKNQDLD SYMEGPSTRL 1200 RVRNLKPSRG SSETKPKKNG KRGSKISFFG VASEDDVEED EEEENEEPST RLRVRIMKSS 1260 RGSSETKPKK TCKKGSSLSF SGVASEDDVE ENEAEENEEE ECAAFQCDME GCTMSFNSEK 1320 QLSLHKRNIC PVKGCGKNFF SHKYLVQHRR VHSDDRPLKC PWKGCKMTFK WAWSRTEHIR 1380 VHTGARPYVC AEPRCGQTFR FVSDFSRHKR KTGHSVKKTK KR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6a57_A | 2e-77 | 1302 | 1422 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a58_A | 2e-77 | 1302 | 1422 | 20 | 140 | Lysine-specific demethylase REF6 |
| 6a59_A | 2e-77 | 1302 | 1422 | 20 | 140 | Lysine-specific demethylase REF6 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 963 | 968 | RKRKRR |
| 2 | 964 | 976 | KRKRRAKTVPRKK |
| 3 | 965 | 976 | RKRRAKTVPRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
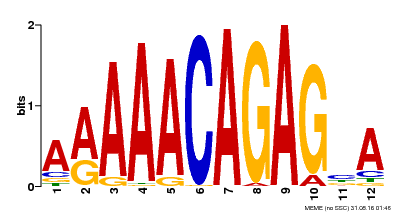
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676776926 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006404261.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A1J3JIY4 | 0.0 | A0A1J3JIY4_NOCCA; Lysine-specific demethylase REF6 | ||||
| STRING | XP_006404261.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



