 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 676781692 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Sisymbrieae; Sisymbrium
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 960aa MW: 107318 Da PI: 5.1875 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 59 | 1.1e-18 | 37 | 83 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg WT+eEde+l++av+ + g++Wk+Ia+++ Rt+ qc +rwqk+l
676781692 37 RGQWTAEEDEILKKAVHSFKGKNWKKIAEYFK-DRTDVQCLHRWQKVL 83
89*****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 62.8 | 6.9e-20 | 89 | 135 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEde +v++++++G++ W+tIar ++ gR +kqc++rw+++l
676781692 89 KGPWTKEEDEMIVQLIQKYGPKKWSTIARFLP-GRIGKQCRERWHNHL 135
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 43 | 1e-13 | 141 | 183 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++ +G++ W+ + ++ gR+++ +k++w+
676781692 141 KEAWTQEEELVLIRAHQIYGNR-WAELTKFLP-GRSDNGIKNHWH 183
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 19.096 | 32 | 83 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.44E-15 | 34 | 89 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.2E-16 | 36 | 85 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.0E-16 | 37 | 83 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 7.1E-23 | 39 | 91 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.86E-14 | 40 | 83 | No hit | No description |
| PROSITE profile | PS51294 | 32.297 | 84 | 139 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.2E-31 | 86 | 182 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.1E-19 | 88 | 137 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-17 | 89 | 135 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.91E-17 | 91 | 135 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-27 | 92 | 139 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.6E-19 | 140 | 190 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-12 | 140 | 188 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.094 | 140 | 190 | IPR017930 | Myb domain |
| Pfam | PF00249 | 9.6E-12 | 141 | 183 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.02E-9 | 143 | 183 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 960 aa Download sequence Send to blast |
MESECAATIS TPPVGIPEAN PKSRHGRTSG PARRSTRGQW TAEEDEILKK AVHSFKGKNW 60 KKIAEYFKDR TDVQCLHRWQ KVLNPELVKG PWTKEEDEMI VQLIQKYGPK KWSTIARFLP 120 GRIGKQCRER WHNHLNPAIN KEAWTQEEEL VLIRAHQIYG NRWAELTKFL PGRSDNGIKN 180 HWHSSVKKKL DSYMSSGLLD QYQAMPLAPY DKSTALQSAF MRSTMDGIGC VSGREEKEIE 240 NCQISSMAGC SISARDYQNG MMNMTQQFHP CENSNKNERA AYHSDQYYYP ELEDISVSIS 300 EASYDMEDCS QFPDHNVSAS SSQDYQFDFQ DLSDISLEMR HNMSELPMPY TKESKESSLG 360 APNSTSNIDV ATCTNPANVS TSETECCRVL FLDQESEGLS VSKSSTQEPN EVDQADYQDS 420 FVCASASESQ TSEATKSPMQ SSSSKSIATA ASGKETLRPA PLIISPEKYS KKSSGLICHP 480 FEAEPNCQTN ENGSFICISD PSTSTCVDEG TNNSSEDQSY HLNDSKKLVP VNDFASLAEV 540 NPHSLPKHEP NMSSEQHHED MGASSSLCFP SLDLPAFNDP LHDYSPLGIR KLLMSTMTCM 600 SPLRLWESPT GKKTLVGAKS VLRKRTRDLL TPLSEKRSDK KLEIDIAASL AKDFSRLDVM 660 FDESESRESI SGTNTGVNLG DKENHFQNLN GEDEESWRGK PTLLSSHRMP EETMDIRKSL 720 KNPEQTCLEA KVRQKDDSEP DVEKAESFSG VLSENNTNKQ VLSPPGQSVT KAEKAQVSTP 780 RNHLQRTLMA TSNKEQHASA SLCLVINSPS RARNSESHLV NNETGNENFS IFCGTPFRRG 840 LESPSAWKSP FYVNSLLPSP RFDTEITIED MGYIFSPGER SYESIGMMTQ RNEHASAFAA 900 FEAMEVSFSP SIDDARKMKD LDKENNDPLL AEGRVLDFND CESPTKVTEE VSSYLLKGCR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 7e-69 | 37 | 190 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 7e-69 | 37 | 190 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (PubMed:21862669). Involved in the regulation of cytokinesis, probably via the activation of several G2/M phase-specific genes transcription (e.g. KNOLLE) (PubMed:17287251, PubMed:21862669, PubMed:25806785). Required for the maintenance of diploidy (PubMed:21862669). {ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:21862669, ECO:0000269|PubMed:25806785}.; FUNCTION: Involved in transcription regulation during induced endoreduplication at the powdery mildew (e.g. G.orontii) infection site, thus promoting G.orontii growth and reproduction. {ECO:0000269|PubMed:20018666}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
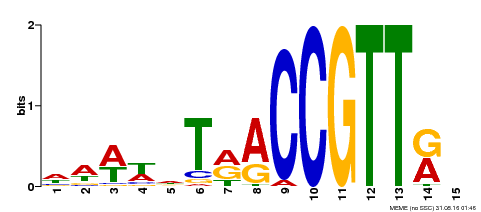
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 676781692 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid (SA) (PubMed:16463103). Expressed in a cell cycle-dependent manner, with highest levels 2 hours before the peak of mitotic index in cells synchronized by aphidicolin. Activated by CYCB1 (PubMed:17287251). Accumulates at powdery mildew (e.g. G.orontii) infected cells. {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:17287251}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF371975 | 0.0 | AF371975.2 Arabidopsis thaliana putative c-myb-like transcription factor MYB3R-4 (MYB3R4) mRNA, complete cds. | |||
| GenBank | AY519650 | 0.0 | AY519650.1 Arabidopsis thaliana MYB transcription factor (At5g11510) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006399657.1 | 0.0 | transcription factor MYB3R-4 | ||||
| Swissprot | Q94FL9 | 0.0 | MB3R4_ARATH; Transcription factor MYB3R-4 | ||||
| TrEMBL | V4KT42 | 0.0 | V4KT42_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006399657.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13213 | 18 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 0.0 | myb domain protein 3r-4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



