 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678397124 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 223aa MW: 24804.1 Da PI: 5.5097 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 82.3 | 3.2e-26 | 9 | 59 | 1 | 51 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
k+i+n+ rqvtfskRr g++KKAeEL vLCd ev +i+fs +gklyey+s
678397124 9 KKIDNTAARQVTFSKRRRGLFKKAEELAVLCDSEVGLIVFSASGKLYEYAS 59
68***********************************************86 PP
| |||||||
| 2 | K-box | 50.1 | 1.2e-17 | 92 | 171 | 19 | 98 |
K-box 19 qelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrkkl 98
++++L ke+ r +R + Ge+L+ Ls++eLqqLe+ +e++l+++ +kK e ++++i +lq+k k+l +en++L++ +
678397124 92 SNYSRLSKEVADRTRHLRLMRGEELQGLSIEELQQLETSIESGLNRVMEKKGEKIMSEIGRLQEKGKQLMDENEKLNQLV 171
67899999999999**************************************************************9866 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 3.1E-37 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 29.927 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 9.42E-39 | 2 | 68 | No hit | No description |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.7E-26 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| SuperFamily | SSF55455 | 1.23E-28 | 3 | 69 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.1E-24 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.7E-26 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 1.7E-26 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS51297 | 11.933 | 87 | 180 | IPR002487 | Transcription factor, K-box |
| Pfam | PF01486 | 1.3E-15 | 92 | 169 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009266 | Biological Process | response to temperature stimulus | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010076 | Biological Process | maintenance of floral meristem identity | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048438 | Biological Process | floral whorl development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000900 | Molecular Function | translation repressor activity, nucleic acid binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 223 aa Download sequence Send to blast |
MAREKIEIKK IDNTAARQVT FSKRRRGLFK KAEELAVLCD SEVGLIVFSA SGKLYEYASS 60 SMKEILERRD LNSKNLTANG RPPVGMQLAE DSNYSRLSKE VADRTRHLRL MRGEELQGLS 120 IEELQQLETS IESGLNRVME KKGEKIMSEI GRLQEKGKQL MDENEKLNQL VADIAYYGND 180 MVAAESMNMG SDEGPWSESL SSSSNGPPQG CHSSDASLKL GLP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6byy_A | 4e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6byy_B | 4e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6byy_C | 4e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6byy_D | 4e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6bz1_A | 5e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6bz1_B | 5e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6bz1_C | 5e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| 6bz1_D | 5e-19 | 1 | 66 | 1 | 66 | MEF2 CHIMERA |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor that coordinates gene expression underlying the differentiation of the pedicel abscission zone. May also be involved in the maintenance of the inflorescence meristem state. | |||||
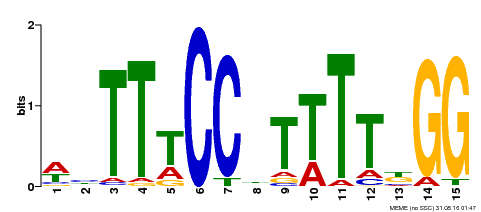
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00272 | DAP | Transfer from AT2G22540 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678397124 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020554864.1 | 1e-97 | MADS-box protein SVP | ||||
| Swissprot | Q9FUY6 | 9e-89 | JOIN_SOLLC; MADS-box protein JOINTLESS | ||||
| TrEMBL | S8D1G3 | 1e-102 | S8D1G3_9LAMI; Uncharacterized protein | ||||
| STRING | XP_009783886.1 | 3e-95 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA938 | 23 | 77 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22540.1 | 1e-83 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



