 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678417860 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 242aa MW: 28127.6 Da PI: 6.9746 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.7 | 2e-16 | 14 | 67 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT......TTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg......kgRtlkqcksrwqkyl 48
rg+W+ eEde+l++++k++G g W++ ++ g + R++k+c++rw +yl
678417860 14 RGKWSVEEDERLINYIKEHGEGGWRLLPKNAGiirfigLLRCGKSCRLRWINYL 67
89**************************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 59.4 | 8.1e-19 | 73 | 118 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T++E+el+++++k lG++ W++Ia++++ Rt++++k++w+++l
678417860 73 RGNFTQDEEELIIELHKILGTK-WSLIAKHLP-WRTDNDIKNYWNSHL 118
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 8.7E-22 | 5 | 70 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.559 | 9 | 67 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.98E-28 | 11 | 114 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-11 | 13 | 69 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-14 | 14 | 67 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.73E-8 | 16 | 67 | No hit | No description |
| PROSITE profile | PS51294 | 26.039 | 68 | 122 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.1E-25 | 71 | 122 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.0E-17 | 72 | 120 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.5E-17 | 73 | 118 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.52E-11 | 75 | 118 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0051555 | Biological Process | flavonol biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 242 aa Download sequence Send to blast |
MGRKPCCEKT GLKRGKWSVE EDERLINYIK EHGEGGWRLL PKNAGIIRFI GLLRCGKSCR 60 LRWINYLRSD VKRGNFTQDE EELIIELHKI LGTKWSLIAK HLPWRTDNDI KNYWNSHLSR 120 KVYRFRPNSS SISKSSFHVS DMKQEMQFGN ATTAATNNTD SFACADCCWS VEQQFDQDTL 180 FMQLEQNGEN NSTRNGNFSA WEPEDFTQFE CYLDVGNQQL GATTGYDQEL WSNQLAMVCN 240 WP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 6e-26 | 12 | 122 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor postulated to regulate the biosynthetic pathway of a flavonoid-derived pigment in certain floral tissues. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00598 | PBM | Transfer from AT5G49330 | Download |
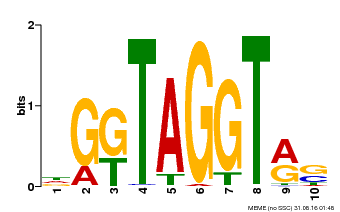
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678417860 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011071652.1 | 4e-65 | transcription factor MYB14 | ||||
| Swissprot | P27898 | 3e-61 | MYBP_MAIZE; Myb-related protein P | ||||
| TrEMBL | A0A0S2SZZ7 | 4e-63 | A0A0S2SZZ7_9LAMI; Light areas 1 | ||||
| TrEMBL | A0A0S2T0F2 | 3e-63 | A0A0S2T0F2_ERYLE; Light areas 1 | ||||
| STRING | Migut.N00057.1.p | 7e-64 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12 | 24 | 2154 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G49330.1 | 8e-56 | myb domain protein 111 | ||||



