 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678447540 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 652aa MW: 72558.4 Da PI: 6.6799 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 404.3 | 1.5e-123 | 36 | 370 | 1 | 320 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkekve 100
eel krmwkd ++l+r+ker+k + +k+ ++ ks+ +++qa rkkm+raQDgiLkYMlk mevc+a+GfvYgiipekgkpv+gasd+LraWWkekv+
678447540 36 EELRKRMWKDHVKLRRIKEREKLVI-QKS--SEKLKSKSTSDQACRKKMARAQDGILKYMLKLMEVCKARGFVYGIIPEKGKPVSGASDNLRAWWKEKVK 132
8*********************877.333..47888999************************************************************* PP
XXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----GGG CS
EIN3 101 fdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkphd 200
fd+ng aai+ky+a++++ ++++++ + ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekg +pPWWPtG+e+ww +lgl++ q ppykkphd
678447540 133 FDKNGLAAIAKYEAECVLKEDGDDN---NGNSQNVLKDLQDATLGSLLSSLMQHCDPPQRKYPLEKGNPPPWWPTGNEEWWMRLGLPTGQT-PPYKKPHD 228
************9999887777777...899********************************************************9998.******** PP
--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX..XX.......XXXXXXXXXXXXXXXXXXXX...... CS
EIN3 201 lkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah..ss.......slrkqspkvtlsceqkedve...... 285
lkk+wk++vLtavikhmsp+i+++r+l rqsk+lqdkm+ake++++lsvl++ee+++ + s + ss ++r +++k t+++++++ ve
678447540 229 LKKMWKIGVLTAVIKHMSPDIAKMRRLIRQSKCLQDKMTAKETAVWLSVLSREEALIWQPSCEngSSgisdapsKSRGHKKKPTVESDSDYVVEdvdsgl 328
*********************************************************9999995533677777656668888999999998777766777 PP
....XXXXXX.X.XXXXXXXXX.......XXXXXXXXXXXXXXXXXX CS
EIN3 286 ....gkkeskik.hvqavktta.......gfpvvrkrkkkpsesakv 320
+s+ + ++q+ + + + v+k +++p++++
678447540 329 gavsS--KSEGRhQSQSMNPSEasscattQ---VKKHDERPRKRKHL 370
66431..22222022222222244442222...22222222222222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 1.7E-121 | 36 | 282 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 8.5E-69 | 158 | 287 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 1.29E-59 | 163 | 286 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 652 aa Download sequence Send to blast |
MEDAEIGISS DIEVDEIRGD EGLEEKDVSD EEIEAEELRK RMWKDHVKLR RIKEREKLVI 60 QKSSEKLKSK STSDQACRKK MARAQDGILK YMLKLMEVCK ARGFVYGIIP EKGKPVSGAS 120 DNLRAWWKEK VKFDKNGLAA IAKYEAECVL KEDGDDNNGN SQNVLKDLQD ATLGSLLSSL 180 MQHCDPPQRK YPLEKGNPPP WWPTGNEEWW MRLGLPTGQT PPYKKPHDLK KMWKIGVLTA 240 VIKHMSPDIA KMRRLIRQSK CLQDKMTAKE TAVWLSVLSR EEALIWQPSC ENGSSGISDA 300 PSKSRGHKKK PTVESDSDYV VEDVDSGLGA VSSKSEGRHQ SQSMNPSEAS SCATTQVKKH 360 DERPRKRKHL KSNAEHAMHG ASRNKHARDD LDGSSMQEHQ NSIPVHGYMM SNSLDEVNRM 420 PSIMLHRNEQ GQANLEESNV ATHCSFPTTN PVAIATADLS NRHLHFPTVQ GSDVIPTGPD 480 TTTQMPQSNN LHGQRQHSQS LGQNTVLDQG LQSSRLHYEP MHVNSPWKYG LQNLAVHPGT 540 QSSSNHQFSS SVAGASDIVT GLSAEVLTDF PYGHEDIATP GSFPHYVKGA LPKEPDRPTG 600 NQFPSPLRTL SPDLTGFSTF DLPFGGDSLL GIDFPVDVDP TDVDYLMEYF AS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 7e-71 | 162 | 285 | 9 | 132 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
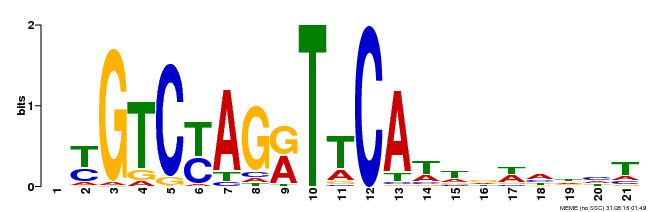
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678447540 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011090401.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-159 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A022PP09 | 0.0 | A0A022PP09_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.N02294.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA304 | 23 | 178 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-152 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



