 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678457497 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1066aa MW: 117749 Da PI: 6.7439 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 71.5 | 1.5e-22 | 151 | 228 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE.-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrf.CqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cqv+gC+++ls ak+yhrrhk v ++ ++ s fh l+efD +krsCrrrLa+hn+rrrk +
678457497 151 VCQVDGCKENLSAAKDYHRRHKAAISIFYDVLVKLKMQMSLLnLWLSSMFHLLREFDAGKRSCRRRLAGHNRRRRKIH 228
6*********************987766666666555544441456899**************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.7E-13 | 146 | 214 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 18.572 | 149 | 227 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.45E-18 | 150 | 229 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.2E-11 | 152 | 226 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 4.4E-4 | 815 | 960 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 6.37E-5 | 815 | 957 | No hit | No description |
| PROSITE profile | PS50297 | 8.587 | 898 | 968 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1066 aa Download sequence Send to blast |
MEEAGGQVIT PIADYQNLVG RFCDSYPTSK KRGVSVHAPN AAYPDTSETW NANSWDWDPT 60 RFVAKPLQCD DVIVVGGQGI LRRNDDQSND SNPRMVHPTR EDQENIFLKL GGDSSAKSGT 120 SGETDSEELQ PSSKPVKRVR AGSPGGANYP VCQVDGCKEN LSAAKDYHRR HKAAISIFYD 180 VLVKLKMQMS LLNLWLSSMF HLLREFDAGK RSCRRRLAGH NRRRRKIHSE DSNTQLLAPQ 240 NRDKNISDVD LVNLLAMLSQ AQHNNRDTNR SFPNKDQLAQ IIGNMNSSLP ETLGPKLKGC 300 VPRHVNSVNQ NSTDKENTSL PSTRDLLAVL TATSPAPRTH DSQSDQTSAE GSESEKSKSS 360 CVNEVAHLNF HSGVTREFPF VGQSSRITSS PPLGDGELNV IETPPSLPLQ LFSSSHDDQH 420 LRKGQDCNVL SAQFKNPSRE AFQLSSKLVV HDFFAMHSSR ENPKDGNLPS GEVELPYLRG 480 NTSNFCSTSL QLYGGPSRLG ENVSFQESHN HMEQPSTSGS YYSTALKSDC QDQTGRIIFK 540 LFDKDPSHLP GSLRTQIYNW LSNSPSDMES YIRPGCILLS VYVAMSPIEW DKLEENLLNY 600 LKMLVDVIDS RFWGAGRFLV HTNRQMAISN GGRIRLCKSG RAGSVPQLIS VSPLAVVSTE 660 ETSILLKGTR LLASSTMIHC THAMGYCIEK HTSSSQNSMY DEILLVGFKV EGASTNRLGR 720 CFIEVENGSR AASFPIIIAD KAICQELKFL EPEIDCRLEI GDGIPTYVAE DEDRSDSREW 780 ALHFLNELGW LFQRKNNSSP SAFQDYSLTR LKFLLMFSVE HDFCAVVKTL LDILAEMNLC 840 QEGLVGDSLE MLSEIQLLTR AVRRRCQGMV NKLIHYSIID PVDNSKKYIF RSDVAGPGGI 900 TPLHLAASLP NAASIVDALT CDPLEVGLRS WGSVLDEQGL SPLEYASVSN NHSYNSLVAQ 960 KLADKSNDQA SISIDNAEQL SVEIDGHKNP SNLRPGTQPC SKCAVVAALG YSRKISGPPA 1020 LLHRSYIHSI LVVAAVCVCV CLLLRGRPFV GYSPPFAWET LDYGAF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-15 | 142 | 226 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00664 | PBM | Transfer from Potri.002G002400 | Download |
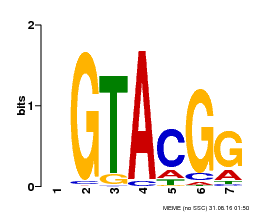
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678457497 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011083361.1 | 0.0 | squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A2G9GTF7 | 0.0 | A0A2G9GTF7_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.C00339.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA12887 | 12 | 19 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76580.1 | 0.0 | SBP family protein | ||||



