 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678459845 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 610aa MW: 67147.6 Da PI: 5.3484 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 53.7 | 3.7e-17 | 250 | 299 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f++Lre++P++ +K +Ka+ L +++eYI+ Lq
678459845 250 RSKHSATEQRRRSKINDRFQKLREVIPNS----EQKKDKASFLLEVIEYIQFLQ 299
899*************************8....7999****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.83E-18 | 245 | 317 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-22 | 245 | 321 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.812 | 248 | 298 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.12E-14 | 250 | 302 | No hit | No description |
| Pfam | PF00010 | 1.5E-14 | 250 | 299 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.7E-12 | 254 | 304 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 610 aa Download sequence Send to blast |
MELPQSRPLE TKGGKATHDF LSLYASSPSN QDPTPPQGSF VVTHDFLQPL ERVGPTDRED 60 NFAGMAAVGN RPPWPAECGT MDPLLPGAIG GYNISYIGQR VSETERNGML AARTSSAHEN 120 DESIFRRWDE YAIVKGKTGK ENAATSRSNL PGMDMVGGEW TSSVLERTSW SPSKQKNTIA 180 PLSSVSSLPP LPSKKKQSFI EIITAAKSSQ GEDEDDMVIK SDTPSHSPLK GSVSLKLENK 240 ALNQKSNTIR SKHSATEQRR RSKINDRFQK LREVIPNSEQ KKDKASFLLE VIEYIQFLQD 300 RVNRYESPYN IWKHEPAKMM QWKNDHIANK QGADTDSGPV LAPAMTFDGN KAVLSPGIPS 360 IGQNLSDSDL TTAKTKEGGQ LVPASALNGT ESPTTINAFS YVAAATSSDA DKTMKWNPLQ 420 SQQMRSSGPE DMISMVDRKM NDHQGQQSTE SGTASISSIY SQGLVNNIAQ ALQRSGINLS 480 QSRVSVKINL GKRANGSIHC STSTAKENHV AAKSTGEDMR FASKKQKRKA EDIDTHMEYE 540 SDFEFDYGSE EKDYEILDDQ FPIENDDYGQ ALEIPMSEGI QSEAPQNETS EGPLNIIRME 600 SDSVRIQELV |
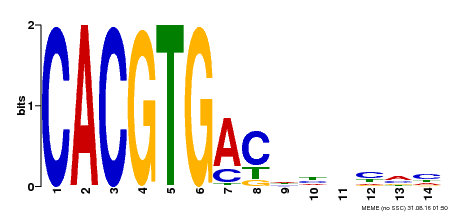
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678459845 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011078688.1 | 1e-145 | transcription factor BIM1 isoform X2 | ||||
| TrEMBL | A0A2G9G0J5 | 1e-147 | A0A2G9G0J5_9LAMI; Uncharacterized protein | ||||
| STRING | Migut.D01611.1.p | 1e-120 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA3969 | 23 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 2e-66 | bHLH family protein | ||||



