 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678465621 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 683aa MW: 74745.6 Da PI: 6.3945 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.6 | 6.9e-16 | 470 | 516 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k+Lq
678465621 470 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLGEAIEYLKMLQ 516
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 4.37E-17 | 462 | 520 | No hit | No description |
| SuperFamily | SSF47459 | 3.79E-20 | 463 | 525 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.746 | 466 | 515 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.1E-13 | 470 | 516 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 9.6E-20 | 470 | 524 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 9.4E-18 | 472 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 683 aa Download sequence Send to blast |
MCRCRLSAGL CETYKAWELS LYSYQPRFVG GEFKEAYSLR PEENEMELMA VSFHGWKRGS 60 FQEKCSWVPA EVRKMPQGEY SRIAGRRHES GNYFPFLNGF SSRVGGDLSE LVWENGQVTV 120 QGQSSKLSRP SLVTSLPSGV PSEPIEGQKI GILHDISPVV PPSELDLCLT NELAPWLDYP 180 FDDGMEQGFG SQRLPEVTAV MEHGISDGNG CELGQSNLDS GEKGPPFSYD PLWPLQNRQA 240 PGGSGSRAQD SISNQTEYLF ENQDQNRDLF SDDRHRRSNS KMVNFPHFSK PNAANLQDPV 300 GIPASGGPNS KNIDTSFHVS AAPEEVSNAM QELAKQVLNE DLPKETDNGN MKLQVQGSET 360 LEKQISCDEL PVEQNVACSS VGSGHGAEVV SSDQTNQCKR KSFDADDSGV HSDDDDIIYV 420 AEKKHAPSGR GSVSRRSEVV EVDTDHVDGK KPTPPRKGTS GSKRSRVAEV HNLSERRRRD 480 RINEKMRALQ ELIPNCNKAD KASMLGEAIE YLKMLQFQVQ FMSMGGGLCM RPVMFPQGIQ 540 PMYHHHHLAP FPPMGVGMSP GFGMGMVDSR SPIFPATLPP MPHIPSPVSG PSSMFRVTGP 600 GLPVYGHPCE GPPAQMNSQH AAVPATGYHN KQPCPPAALN PTNPNTKTSS RSASSAAVSR 660 RSRQVEAMNQ STTTQDVNAV HSK |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 474 | 479 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
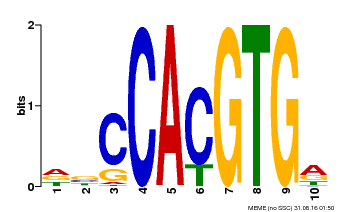
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678465621 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 6e-40 | phytochrome interacting factor 3 | ||||



