 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 678466846 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Lamiales; Lentibulariaceae; Utricularia
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 821aa MW: 94426.2 Da PI: 7.1905 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 88.8 | 7.8e-28 | 64 | 151 | 1 | 91 |
FAR1 1 kfYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkktekerrtraetrtgCkaklkvkkekdgkwevtkleleHnHelap 91
+fY+eYAk++GF++++++s++sk+++e+++++f Cs++g + e++++ ++r++++++t+Cka+++vk+++dg w+++++ +eHnHel p
678466846 64 SFYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYGMTPESETS---SSRRSSVKKTDCKASMHVKRKRDGGWYIHEFIKEHNHELLP 151
5***************************************9998877...889999********************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.8E-25 | 64 | 151 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.9E-20 | 272 | 364 | IPR018289 | MULE transposase domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010018 | Biological Process | far-red light signaling pathway | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 821 aa Download sequence Send to blast |
MVGCEGNAKC GRGDALNVID AVDSVQSRDD PLNTCPKTDV SGIEGSRFEP DEGIDFESHE 60 AAYSFYQEYA KSMGFTTSIK NSRRSKKTKE FIDAKFACSR YGMTPESETS SSRRSSVKKT 120 DCKASMHVKR KRDGGWYIHE FIKEHNHELL PALAYHFHIH RNVRLAEKNN IDILHAVSER 180 TRKMYVEMCR QYGGIHEPSF SKNEVDHQAD GARCMALDEG DARFMMDTFM QLQKESPYFS 240 YAVELNREQR VRNLFWIDSK SRKDYIFFSD AVFLDTTFVK SSDKVPIALF LGVNHHFQPM 300 LLGCALLADE SKSTFVWLLK TWLRAVGGKA PNLILSCQDT QLKLAIEEIF PCSRHCFPLW 360 HVMEKIPVTL GHVLRLHVNF IKKFNKCISR SLTDEEFDMR WWKMVTRFEL QENEWVHSLY 420 VERKKWAPIF MRDAFLAGLS PRQRSDSVNS FFDKYIHKKM NVNEFMRQYI VILQNRSEEE 480 DLADFDMLHR QPALKSPSPW EKQMSTIYTH SIFRKFQVEV LGVVGCHPKK EREDGVNVTF 540 KVDDCEKGEN FVVMWNEEMS ELCGLSRIPS QYILKRWTKD AKVRQGDLEE TDSAHTRVQR 600 YNDLCKIAIA LGEEGSSCEE NYNIACRALV QALKNCVNVN NRTAVDSSSN SVGLRFSEQE 660 TLLLHGNKSS KKKNSGKKRK QEPEPIVNET EDNIEPLETL SSQEMTLNGY YGSQQNVHGM 720 LNLMEPPQDG YYVSQPTLPG MVRRSACGVA CALMFHVTCF IKDECHYSQL NSIPSGHDSF 780 YGAQQNLPAL GHLDFRQATF PYIMQGDHSV RQPELQEGAR N |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00434 | DAP | Transfer from AT4G15090 | Download |
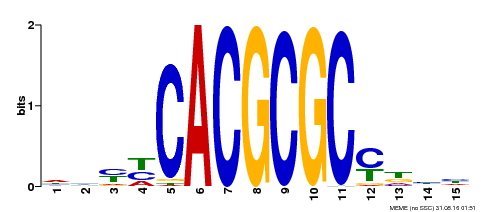
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 678466846 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. {ECO:0000269|PubMed:18033885}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011085456.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| Refseq | XP_011085457.1 | 0.0 | protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| Swissprot | Q9SWG3 | 0.0 | FAR1_ARATH; Protein FAR-RED IMPAIRED RESPONSE 1 | ||||
| TrEMBL | A0A022RW20 | 0.0 | A0A022RW20_ERYGU; Uncharacterized protein | ||||
| STRING | Migut.H01263.1.p | 0.0 | (Erythranthe guttata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA496 | 21 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G15090.1 | 0.0 | FAR1 family protein | ||||



