 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 891847 | ||||||||
| Common Name | ARALYDRAFT_891847 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 227aa MW: 25855.2 Da PI: 9.6485 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 53.1 | 5.7e-17 | 129 | 222 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE..EEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefelvvkvfrk 99
f vl ps+ + ++++lp fae++ +g++ + ++ ++ ++W v++ k gr +++GW eF +n++ egD++vF+l ++++f l+v++fr
891847 129 FRVVLRPSYLYRGCIMYLPSGFAEKYlSGIS-GF--IK-LQLGEKQWPVRC--LYKAGRAKFSQGWYEFTLENNIGEGDVCVFELLRTRDFILKVTAFRV 222
678899********************85452.33..44.44688*******..666666779************************99***9*9999985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 3.0E-23 | 121 | 221 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 1.71E-22 | 122 | 223 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 4.04E-21 | 127 | 221 | No hit | No description |
| PROSITE profile | PS50863 | 12.52 | 129 | 223 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.6E-19 | 129 | 223 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.0E-15 | 129 | 222 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0034613 | Biological Process | cellular protein localization | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 227 aa Download sequence Send to blast |
MEMDSAQNQF NKRARLFEDS QDKDAKVIYP SNPESTEPVN KGYGGSTAIQ SFFKESKAEE 60 TPKVLKKRGR KKKNPNPEEV NSSTPGGDDS ENRSKFYESA SARKRTVTAE ERERAVNAAK 120 TFEPTNPYFR VVLRPSYLYR GCIMYLPSGF AEKYLSGISG FIKLQLGEKQ WPVRCLYKAG 180 RAKFSQGWYE FTLENNIGEG DVCVFELLRT RDFILKVTAF RVNEYV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4i1k_A | 3e-92 | 95 | 226 | 15 | 146 | B3 domain-containing transcription factor VRN1 |
| 4i1k_B | 3e-92 | 95 | 226 | 15 | 146 | B3 domain-containing transcription factor VRN1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the regulation of vernalization (By similarity). Binds to DNA in vitro in a non-sequence-specific manner. XLG2 promotes the DNA binding activity of RTV1 specifically to promoter regions of FT and SOC1 in vivo and thus leads to the activation of floral integrator genes. {ECO:0000250, ECO:0000269|PubMed:22232549}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00192 | DAP | Transfer from AT1G49480 | Download |
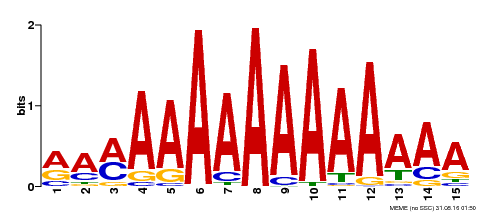
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 891847 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cold. {ECO:0000269|PubMed:22232549}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | BT015682 | 0.0 | BT015682.1 Arabidopsis thaliana At1g49480 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002894192.1 | 1e-167 | B3 domain-containing protein REM19 | ||||
| Swissprot | Q9XIB5 | 1e-164 | REM19_ARATH; B3 domain-containing protein REM19 | ||||
| TrEMBL | D7KEX6 | 1e-166 | D7KEX6_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_104641.1 | 1e-166 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1124 | 28 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49480.1 | 1e-155 | related to vernalization1 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 891847 |
| Entrez Gene | 9327576 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



