 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | 907523 | ||||||||
| Common Name | ARALYDRAFT_907523 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Arabidopsis
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 922aa MW: 103248 Da PI: 5.7552 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 134.1 | 4.5e-42 | 121 | 197 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
Cqve+C +dls++k+yhrrhkvCe+hska+++lv g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
907523 121 CQVENCGVDLSKVKDYHRRHKVCEMHSKATSALVGGIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANP 197
**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.0E-34 | 116 | 182 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.496 | 118 | 195 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 6.93E-40 | 120 | 200 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.1E-31 | 121 | 194 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 3.4E-6 | 707 | 812 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 1.24E-6 | 709 | 810 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 8.79E-8 | 712 | 810 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 922 aa Download sequence Send to blast |
MEARIEGEEE GHSLEYGFSG KRSVEWDLND WKWNGDLFVA TQLNHGSSNS SSTCSDEAIV 60 EIMEKKKKRG AVTVIAMEED NLRDGDDDAH RLTLNLGGNI EGNGAKKTKL GGGIPSRAIS 120 CQVENCGVDL SKVKDYHRRH KVCEMHSKAT SALVGGIMQR FCQQCSRFHV LEEFDEGKRS 180 CRRRLAGHNK RRRKANPDTI GNGNPLSDGQ TSNYLLITLL KILSNLHSNQ SDQTGDQDLL 240 SQLLKSLVSQ AGEHIGKNLV GLLQGGGGVQ ASQNIGNSSD LLSLEQAPRE NIKHHSVPET 300 HWQEVFANGA QETAAPDRPE KQVKMNDFDL NDIYIDSDDT TDIERSSAPP TNPATSSLDY 360 HQDSRQSSPP QTSRRNSDSA SDQSPSSSGG DAQSRTDRIV FKLFGKEPND FPVALRGQIF 420 NWLAHTPTDM ESYIRPGCIV LTIYLRQDEA SWEELCCDLS FSLRRLLDLS DDPLWTDGWI 480 YLRVQNQLAF AFDGQVVLDT SLPLRSHDYS QIITVRPLAV TRKAQFTVKG INLRRHGTRL 540 LCAVEGTYLV QEATQGVMEE RDDLRVNNEI DFVKFSCEMP IASGRGFMEI EDQGGLSSSY 600 FPFIVSEDED VCSEIRRLES TLEFTGTDSA MQAMDFIHEI GWLLHRSELK SRLAASDHNP 660 EDLFPLIRFK FLVEYSMDRE WYAVIKKLLN ILFEEGTVDP SPDAALSELC LLHRAVRKNS 720 KPMVEMLLRF IPKKKNQTSS GLFRPDAAGP GGLTPLHIAA GKDGSEDVLD ALTEDPEMIG 780 IQAWKISQDN TGFTPEDYAR LRGHFSYIHL VQRKLNRKPT AEEHVVVNIP ESFNIEHKQE 840 KRSLMDSSSL SISQINQCKL CDHKRVFVTT QHKSLAYRPA MLSMVAIAAV CVCVALLFKS 900 CPEVLYVFQP FRWELLEYGT S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj0_A | 7e-33 | 118 | 175 | 3 | 60 | squamosa promoter-binding protein-like 12 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00060 | PBM | Transfer from AT3G60030 | Download |
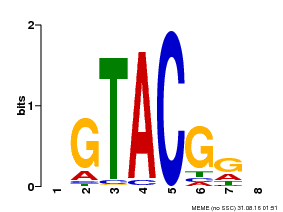
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | 907523 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ132096 | 0.0 | AJ132096.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002878311.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Refseq | XP_020880464.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Refseq | XP_020880465.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | D7LWK1 | 0.0 | D7LWK1_ARALL; Uncharacterized protein | ||||
| STRING | scaffold_503227.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | 907523 |
| Entrez Gene | 9314379 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



