 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA10G00196 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 642aa MW: 73607.3 Da PI: 7.8849 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 166.2 | 1.2e-51 | 10 | 143 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk..kg 97
lppGfrFhPtdeel+ +yL+kkv+++ l+l ++i++vdiyk++PwdLp+k+ +ekewyfFs+rd+ky++g r+nra++sgyWkatg+dk + ++ +
AA10G00196 10 LPPGFRFHPTDEELILHYLRKKVSSSLLPL-SIIADVDIYKFDPWDLPAKAPFGEKEWYFFSPRDRKYPNGARPNRAAASGYWKATGTDKLIATSanN- 106
79****************************.89***************988899***************************************98650. PP
NAM 98 el.......vglkktLvfykgrapkgektdWvmheyrl 128
+ +g+kk Lvfykg+ pkg+kt+W+mheyrl
AA10G00196 107 -NnggfnenIGVKKALVFYKGKPPKGVKTNWIMHEYRL 143
.33455556***************************98 PP
| |||||||
| 2 | NAM | 172 | 1.8e-53 | 339 | 465 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgel 99
lppGfrFhPtdee+ ++yLk+kv + ++++ +i e+d++k ePwdLpkk+k +eke yfF++rd+ky+tg r+nrat sgyWkatgkdke+++ k+ l
AA10G00196 339 LPPGFRFHPTDEEIATHYLKEKVLNPQFTA-AAIGEADLNKNEPWDLPKKAKMGEKEFYFFCQRDRKYPTGMRTNRATVSGYWKATGKDKEIFRGKTCL 436
79*************************999.78***************99999*****************************************99999 PP
NAM 100 vglkktLvfykgrapkgektdWvmheyrl 128
vg+kktLvfykgrapkgekt+Wvmhe+rl
AA10G00196 437 VGMKKTLVFYKGRAPKGEKTNWVMHEFRL 465
***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.44E-60 | 8 | 183 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.873 | 10 | 183 | IPR003441 | NAC domain |
| Pfam | PF02365 | 9.4E-28 | 11 | 143 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 2.75E-60 | 336 | 491 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.352 | 339 | 490 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.7E-27 | 340 | 465 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:1900057 | Biological Process | positive regulation of leaf senescence | ||||
| GO:1903648 | Biological Process | positive regulation of chlorophyll catabolic process | ||||
| GO:0000975 | Molecular Function | regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 642 aa Download sequence Send to blast |
MISKDPRSSL PPGFRFHPTD EELILHYLRK KVSSSLLPLS IIADVDIYKF DPWDLPAKAP 60 FGEKEWYFFS PRDRKYPNGA RPNRAAASGY WKATGTDKLI ATSANNNNGG FNENIGVKKA 120 LVFYKGKPPK GVKTNWIMHE YRLSDSLSPK RHRSNITTNS NNFKSKDYSM RLDDWVLCRI 180 YKKSNASLST PVIAADQEHE EIDQESSILP NLAIDRSLKQ QKSCSFSNLL DAADLTFLTD 240 IFNENNQEIT RNESDFSLML GNFSNYQEDQ FLDQKLPQLS SIPMEKTVIG SKRERLDFDD 300 ETTTNPWKKM MMRVERKLIM MVEEGGVVVN QGSEEVIKLP PGFRFHPTDE EIATHYLKEK 360 VLNPQFTAAA IGEADLNKNE PWDLPKKAKM GEKEFYFFCQ RDRKYPTGMR TNRATVSGYW 420 KATGKDKEIF RGKTCLVGMK KTLVFYKGRA PKGEKTNWVM HEFRLDGKYS YHNLPKSARD 480 EWVVCRVFHK NSPIAPPTQN QVMMRRQPTM EDFSRLDSFE NIDHLLDFSS LPPLMDQPGP 540 SFSNDQHDFK PINPSYFPNQ TFNFPYQTGP MFPREISNKG MVKLEHSFVS VSQDTGLSSD 600 VNTIATPEIS SYQGRTGVAN PAMIDGSKCT YDDLDDLEIL WD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 9e-68 | 9 | 492 | 15 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 9e-68 | 9 | 492 | 15 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 9e-68 | 9 | 492 | 15 | 169 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 9e-68 | 9 | 492 | 15 | 169 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 9e-68 | 9 | 492 | 15 | 169 | NAC domain-containing protein 19 |
| 4dul_B | 9e-68 | 9 | 492 | 15 | 169 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that acts as positive regulator of leaf senescence. Activates NYC1, SGR1, SGR2 and PAO, which are genes involved in chlorophyll catabolic processes. Activates senescence-associated genes, such as RNS1, SAG12 and SAG13. {ECO:0000269|PubMed:27021284}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00332 | DAP | Transfer from AT3G04060 | Download |
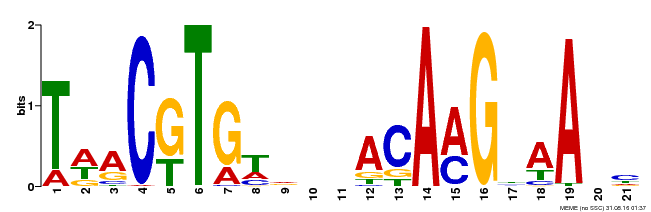
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA10G00196 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010518044.1 | 1e-164 | PREDICTED: NAC domain-containing protein 92-like | ||||
| Swissprot | Q9SQQ6 | 1e-164 | NAC46_ARATH; NAC domain-containing protein 46 | ||||
| TrEMBL | A0A1J3CDP6 | 1e-162 | A0A1J3CDP6_NOCCA; Uncharacterized protein | ||||
| TrEMBL | A0A1J3GPP1 | 1e-162 | A0A1J3GPP1_NOCCA; Uncharacterized protein (Fragment) | ||||
| STRING | XP_010518044.1 | 1e-163 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2269 | 27 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04060.1 | 1e-166 | NAC domain containing protein 46 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



