 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA18G00187 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 891aa MW: 102019 Da PI: 8.0109 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 96.6 | 2e-30 | 29 | 158 | 3 | 117 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvgg...................vevl 81
+e +rwl+++ei+a+L n++++ + ++ + pksg+++L++rk++r+fr+DG++wkkkkdgkt++E+he+LKv e +
AA18G00187 29 EEaYSRWLRPNEIHAVLCNHKHFPINVKPVNLPKSGTVLLFDRKMLRNFRRDGHNWKKKKDGKTIKEAHEHLKVWLvtrkgymytmptvtitlplFEGV 127
55589*********************************************************************9877888888888888888888888 PP
CG-1 82 ycyYahseenptfqrrcywlLeeelekivlvhylev 117
++y+ e++++f +++e+ivlvhy+e+
AA18G00187 128 TGYWISYENKSSFPSN-----GRSQEHIVLVHYRET 158
8888888888888774.....345689999999986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 48.176 | 24 | 164 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.6E-39 | 27 | 159 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.5E-24 | 31 | 102 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.64E-13 | 342 | 428 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 5.01E-14 | 528 | 638 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.7E-16 | 528 | 642 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 7.46E-17 | 537 | 652 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.273 | 546 | 611 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 2.7E-6 | 579 | 608 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.808 | 579 | 611 | IPR002110 | Ankyrin repeat |
| Pfam | PF13637 | 5.9E-6 | 581 | 626 | No hit | No description |
| SuperFamily | SSF52540 | 2.12E-5 | 692 | 793 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 98 | 723 | 745 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.035 | 727 | 751 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0011 | 765 | 787 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.56 | 766 | 790 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.7E-4 | 767 | 787 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 6.1 | 843 | 865 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.59 | 844 | 873 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.23 | 845 | 865 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 891 aa Download sequence Send to blast |
MDGVGGRLAG SEIHGFHTFR DLDVQTMLEE AYSRWLRPNE IHAVLCNHKH FPINVKPVNL 60 PKSGTVLLFD RKMLRNFRRD GHNWKKKKDG KTIKEAHEHL KVWLVTRKGY MYTMPTVTIT 120 LPLFEGVTGY WISYENKSSF PSNGRSQEHI VLVHYRETLE VAPATPGNSY SSSITDNLSP 180 KLVSEDIISG HHNACNTVCS NSLVVRNHEN KLHEINTLDW DDLLVPSDNS NQPAPTQEDM 240 SYFMEQLQPV VRNSANHENN LACYNTPTDI QSFPGLGETA HQNENGLQSE ESFGRWLNSF 300 ISDSPGSVED PSFESMVTHG QDPSATSVIR NSPSNIPEQV FNITDVSPAW AYSVERTKIL 360 ITGFFHDSFQ HYERSNLFCI CGESCVTAEY LQVGVYRCFL PPQSPGVVDL YLSADGHKPI 420 SQFFSFEHRS HPVLEKAVPQ DDELYKWEEF EFQVRLAHLL FTSSNKISAL STKISPVNIQ 480 EAKKLANRTS HLLNSWAYLM KSIQANEVSF DQAKDHLFEL TLKNRLKEWL LEKVLENRNT 540 KEYDSKGLGV IHLCAVLGYT WSIHLFSWAG ISLDFRDKHG WTALHWAAYY GREKMVAALL 600 SAGARPNLVT DPNKQNLGGC TAADLAQEKG YDGLSAFLAE KSLIAQFKDM KMAGNINGNI 660 EAVKTEMLSP ETVNEEEQSL KDTLAAYRTA AEAAARIQGA FREHELKVRS SAVKFGSKED 720 EVKNIIAAMK IQHAFRNYEM RRKITAAARI QFRFQTWKMR REFLNMRKKT IKIQAVFRGF 780 QVRRQYRKIT WSVGVLEKAI LRWRLKRKGF RGLQLSQTEG NQEESSDPVE DFYKTSQKQA 840 EDRLERSVVR VQAMFRSKKA QKDYRRMKMT HQEAQLEYDE LQERDEMAME T |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
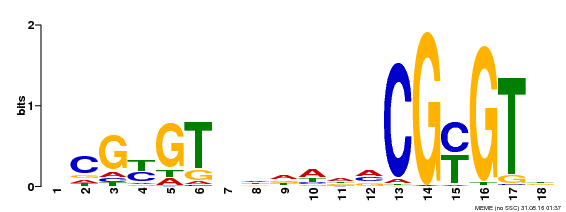
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA18G00187 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006414383.1 | 0.0 | calmodulin-binding transcription activator 5 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | V4P888 | 0.0 | V4P888_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006414383.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4021 | 27 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



