 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA1G00054 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 572aa MW: 63952.2 Da PI: 6.6887 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40 | 7.2e-13 | 412 | 457 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr+++P+ + K++Ka+ L Av YI++L
AA1G00054 412 NHVEAERQRREKLNQRFYALRSVVPNiS------KMDKASLLGDAVSYINEL 457
799***********************66......***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.1E-52 | 48 | 236 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.326 | 408 | 457 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.76E-19 | 411 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.04E-13 | 411 | 462 | No hit | No description |
| Pfam | PF00010 | 1.9E-10 | 412 | 457 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.1E-18 | 412 | 471 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.9E-16 | 414 | 463 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 572 aa Download sequence Send to blast |
MNIGGQIWNE DDKAIVASLL GKRALDYLLS NSVSNANLLM TVGSDEDLQD KLSDLVERPN 60 SSNFSWNYAI FWQISRSKSG DLVLCWGDGS CREPKEGEES EMVKLLSLGR EEETHQKMRK 120 RVLQKLHGLF GGLEEDNCAL GLDRVTDTEM FLLASMYFSF PKGEGGPGKV FLSGKSLWLS 180 DVVNSSSDYC VRSFLAKSAG IKTIVLVPTD IGVVELGSTR SLPENQDSMK SIKSLFSSPR 240 ASLPLVLSEK RDEIGFGVSG FECPPKIFGK DLHQQQQQQQ QHRQFREKLT VRKMEDRVPK 300 RLESAAAAYP NGNRLMFSTS NNNSLINPTW VQPEIYSGRG ITAKEASDEF KFLPLQSQRP 360 PAQMQIDFSG ATSRASENSD GEGAEWVDIT DDGTRPRKRG RRPANGRAEA LNHVEAERQR 420 REKLNQRFYA LRSVVPNISK MDKASLLGDA VSYINELHAK VKVMEAERER LGYSLNPGMM 480 SLEPEINVES ISEDDVVRVR VSCSMESHPA SRMFHVFEEG KVEVIKSNLE VAEERVLHTF 540 VIKSGELTKE KLISAFSREH SSSVQSRTSS GR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 1e-26 | 406 | 468 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
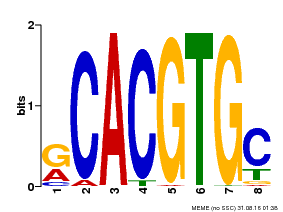
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA1G00054 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001077440.1 | 0.0 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | ||||
| Refseq | NP_001184883.1 | 0.0 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | ||||
| Refseq | NP_171634.1 | 0.0 | basic helix-loop-helix (bHLH) DNA-binding superfamily protein | ||||
| Swissprot | Q9LNJ5 | 0.0 | BH013_ARATH; Transcription factor bHLH13 | ||||
| TrEMBL | A0A178WJW5 | 0.0 | A0A178WJW5_ARATH; JAM2 | ||||
| STRING | AT1G01260.1 | 0.0 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3758 | 27 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



