 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA210G00005 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 444aa MW: 47704.5 Da PI: 8.8777 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 45.3 | 2e-14 | 344 | 392 | 5 | 53 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelk 53
+r+rr++kNRe+A rsR+RK+a++ eLe +v++L++eN++L+ + e+
AA210G00005 344 RRQRRMIKNRESAARSRARKQAYTVELEAEVAKLKEENERLQRKQAEIL 392
79*************************************9976544443 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.20.5.170 | 6.7E-15 | 334 | 403 | No hit | No description |
| SMART | SM00338 | 1.0E-14 | 340 | 405 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 11.346 | 342 | 391 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 4.9E-12 | 344 | 394 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 7.5E-11 | 344 | 393 | No hit | No description |
| CDD | cd14707 | 1.05E-28 | 344 | 398 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 347 | 362 | IPR004827 | Basic-leucine zipper domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009738 | Biological Process | abscisic acid-activated signaling pathway | ||||
| GO:0010255 | Biological Process | glucose mediated signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 444 aa Download sequence Send to blast |
MGSQLNFKNL GNEPPGDGGG GMNRLSQGNT PLTRQGSIYS LTFDEFQSSL GKDFGSLNMD 60 ELLKNIWSAE ETQAMASTSG IPVSGQDGLQ RQGSLTLPRT LSQKTVDEVW KDLSKEYISG 120 KDRGVGGTNM SQTHTQRQQT LGEVTLEEFL VRAGVVREEN QMASNLKPDK GGFFGNPLPA 180 SASPVRFQQA SGVAASLMTN LGPDAANHMQ GQGSSLPLNV NGVRLIDQQH HCVQQQPIMP 240 KQPGFGYGTK IGLPNGQVTN PGIRSGLMGV GDHSLTNNMG LVQGGLGLTG AVGVGAVSPV 300 TPLSSEGIGK SNGDSSSLSP SPYMFNGGVR GRKSSGVVEK VVERRQRRMI KNRESAARSR 360 ARKQAYTVEL EAEVAKLKEE NERLQRKQAE ILEMQKNQLN NYLIANIFKV MNLVVYATGD 420 GDDESSRRAA EKAEEDAVRS LVNE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in ABA and stress responses and acts as a positive component of glucose signal transduction. Functions as transcriptional activator in the ABA-inducible expression of rd29B. Binds specifically to the ABA-responsive element (ABRE) of the rd29B gene promoter. {ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00186 | DAP | Transfer from AT1G45249 | Download |
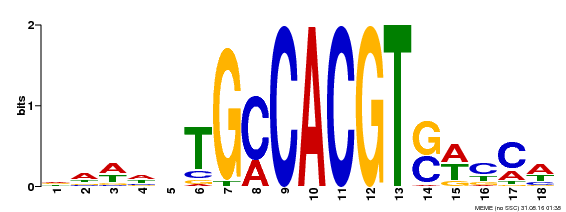
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA210G00005 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by drought, salt, abscisic acid (ABA), cold and glucose. {ECO:0000269|PubMed:10636868, ECO:0000269|PubMed:11005831, ECO:0000269|PubMed:15361142, ECO:0000269|PubMed:16284313, ECO:0000269|PubMed:16463099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010538116.1 | 1e-173 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| Refseq | XP_010538117.1 | 1e-173 | PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| Swissprot | Q9M7Q4 | 1e-166 | AI5L5_ARATH; ABSCISIC ACID-INSENSITIVE 5-like protein 5 | ||||
| TrEMBL | A0A1P8AV75 | 1e-163 | A0A1P8AV75_ARATH; Abscisic acid responsive elements-binding factor 2 | ||||
| STRING | XP_010538116.1 | 1e-172 | (Tarenaya hassleriana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM734 | 27 | 129 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G45249.1 | 1e-122 | abscisic acid responsive elements-binding factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



