 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA26G00745 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 326aa MW: 36010.3 Da PI: 8.1499 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.1 | 1.4e-12 | 257 | 301 | 7 | 55 |
HHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 7 erErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+Er RR +i +++ +L++l+P+ ++ + a++Le Av+YIk+Lq
AA26G00745 257 VAERVRRTKISERMKKLQDLVPNM----DTQTNTADMLESAVQYIKDLQ 301
78*********************7....79******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50888 | 14.856 | 250 | 300 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.22E-16 | 253 | 317 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.09E-11 | 254 | 305 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 5.9E-16 | 255 | 311 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.8E-13 | 256 | 306 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.2E-10 | 257 | 301 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0042335 | Biological Process | cuticle development | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MESYSPQHFH LHQKHQTNSG LTRYQSAPSS YFSTFVNKES IEEFLDRPTS PETEKILSGI 60 LQTTETSKNI DSFLHHSFSR KNNSEENSSS EINVKPLEVK TEEVKTEFEE TALCPGESIG 120 YDSVSKSLNQ KKRQRNNGDL SQTTSLARHN SSPAGLFSSI DVETACAAVM KSMGDIGAVN 180 VMKTNNTEAI PVTPQSKSGF SSRLPPRTLS GSASGRPPLA HHVSLPKSLS DIEEMLSDSI 240 PCKIRAKRGC ATHPRSVAER VRRTKISERM KKLQDLVPNM DTQTNTADML ESAVQYIKDL 300 QEQVKILEEK RGRCSCRPPG PAESGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00195 | DAP | Transfer from AT1G51140 | Download |
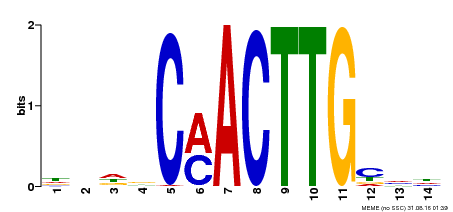
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA26G00745 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_020869674.1 | 1e-139 | transcription factor bHLH122 | ||||
| Swissprot | Q9C690 | 1e-135 | BH122_ARATH; Transcription factor bHLH122 | ||||
| TrEMBL | A0A1J3H6A2 | 1e-139 | A0A1J3H6A2_NOCCA; Uncharacterized protein | ||||
| STRING | fgenesh2_kg.1__4183__AT1G51140.1 | 1e-138 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10547 | 26 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G51140.1 | 1e-132 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



