 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA29G00244 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 344aa MW: 38206.2 Da PI: 4.5874 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 56.7 | 5.9e-18 | 83 | 133 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+ y+GVr++ +g+WvAeIr+p+ r +r +lg++ + +eAa a+++a+k+++g
AA29G00244 83 CNYRGVRQRT-WGKWVAEIREPK----RgTRLWLGTYPSSYEAALAYDEAAKAMYG 133
56*****999.**********83....46************************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 6.6E-14 | 83 | 133 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.45E-31 | 83 | 142 | No hit | No description |
| SuperFamily | SSF54171 | 1.37E-18 | 84 | 140 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.2E-28 | 84 | 142 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 21.074 | 84 | 141 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 4.4E-35 | 84 | 147 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-8 | 85 | 96 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.3E-8 | 107 | 123 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0010286 | Biological Process | heat acclimation | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 344 aa Download sequence Send to blast |
MGVLDQVANL ASIIPFDSSR KRKSRGRRNG ISVAETLRKW KEYNELTEAS CSDGSPTPIR 60 KAPAKGSKKG CMKGKGGPEN GICNYRGVRQ RTWGKWVAEI REPKRGTRLW LGTYPSSYEA 120 ALAYDEAAKA MYGQSARLNQ PHIEDSSCPT AATASGSDTT SEVCRVEDTK MRSNSQVKLE 180 DGSDEYIPLP VPLNSFSDVK EEKGEVKELS SAMASGYELD PKKEMLDGGV VGNGHDQEPW 240 SFDVEEMFDV DELLGSLEEN DVSIGQEKTL QVQDNIGEGD RNMNFNYQMQ FPDANLLGST 300 HLGDDFGYPF VQPSEEQNNG ICFDHRRFQD LDIQDLDFGN THFS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 3e-16 | 84 | 141 | 2 | 60 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 18 | 23 | SRKRKS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]CCGAC-3'. Binding to the C-repeat/DRE element mediates high salinity- and abscisic acid-inducible transcription. {ECO:0000269|PubMed:11798174}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00302 | DAP | Transfer from AT2G40340 | Download |
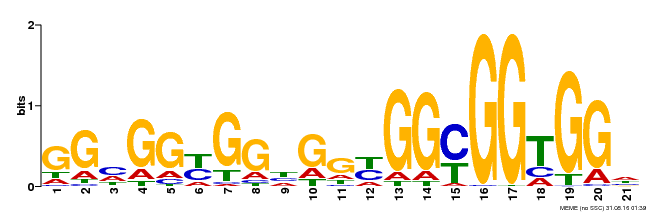
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA29G00244 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By high-salt stress and abscisic acid (ABA) treatment. {ECO:0000269|PubMed:11798174}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006294524.1 | 1e-136 | dehydration-responsive element-binding protein 2C isoform X1 | ||||
| Swissprot | Q8LFR2 | 1e-118 | DRE2C_ARATH; Dehydration-responsive element-binding protein 2C | ||||
| TrEMBL | R0FWR9 | 1e-135 | R0FWR9_9BRAS; Uncharacterized protein | ||||
| STRING | XP_006294524.1 | 1e-136 | (Capsella rubella) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8532 | 25 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40340.1 | 1e-86 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



