 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA30G00231 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 274aa MW: 30231.7 Da PI: 6.3101 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 51.4 | 1.9e-16 | 119 | 165 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L++l+P++ K +Ka++L +A+eY+k+Lq
AA30G00231 119 VHNLSEKRRRSRINEKMKALQSLIPNS-----NKTDKASMLDEAIEYLKQLQ 165
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.92E-17 | 111 | 169 | No hit | No description |
| SuperFamily | SSF47459 | 3.01E-20 | 113 | 175 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.063 | 115 | 164 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 8.6E-20 | 116 | 175 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 6.6E-14 | 119 | 165 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 4.0E-18 | 121 | 170 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MADKKLISSS SSCTYNPPSA SSSDEISLFL RHIFDRSSPL PSYYSTAATA TEINGEQHAE 60 MRSFVSQSSA TRVSSSSIGA SGNETDEYDC ESEEAVVDEP PAKSGPPRSS SKRCRAAEVH 120 NLSEKRRRSR INEKMKALQS LIPNSNKTDK ASMLDEAIEY LKQLQLQVQM LTMRNGLSLH 180 PLCLPGSTLQ LPQIRPVEST NESLINHTNQ FATNPNPPDL INTATSYTLE PAIRNHFGPF 240 PLLNPSMEMN REEGLNLTQQ RLSIGHHNTN LTGE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that plays a role in floral organogenesis. Promotes the growth of carpel margins and of pollen tract tissues derived from them. {ECO:0000269|PubMed:10225997, ECO:0000269|Ref.8}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
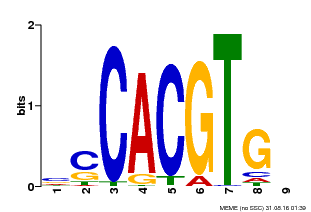
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA30G00231 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by the A class gene AP2 in the first whorl and by ARF3/ETT in gynoecium. {ECO:0000269|PubMed:11245574}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF319540 | 9e-71 | AF319540.1 Arabidopsis thaliana SPATULA (SPT) mRNA, complete cds. | |||
| GenBank | AK229267 | 9e-71 | AK229267.1 Arabidopsis thaliana mRNA for putative bHLH transcription factor (AtbHLH024) / SPATULA (SPT), complete cds, clone: RAFL16-54-P05. | |||
| GenBank | BT026462 | 9e-71 | BT026462.1 Arabidopsis thaliana At4g36930 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019088981.1 | 1e-117 | PREDICTED: transcription factor SPATULA-like isoform X2 | ||||
| Swissprot | Q9FUA4 | 1e-102 | SPT_ARATH; Transcription factor SPATULA | ||||
| TrEMBL | A0A397YC47 | 1e-115 | A0A397YC47_BRACM; Uncharacterized protein | ||||
| STRING | XP_010437251.1 | 1e-116 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4199 | 27 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 6e-92 | bHLH family protein | ||||



