 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA31G00568 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 579aa MW: 65545.3 Da PI: 5.0673 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 432 | 5.7e-132 | 31 | 395 | 1 | 352 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWkekv 99
e+l+krmwkd+++lkr+ker+k ++ +++ +k+++qa+rkkmsraQDgiLkYMlk mevc+++GfvYgiipekgkpv+g+sd++raWWkekv
AA31G00568 31 EDLEKRMWKDRVRLKRIKERQKIGS-----QVTKETPKKISDQAQRKKMSRAQDGILKYMLKLMEVCKVRGFVYGIIPEKGKPVSGSSDNIRAWWKEKV 124
89****************9999632.....35677899************************************************************* PP
XXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-----G CS
EIN3 100 efdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtppykkp 198
+fd+ngpaai+ky+ ++l++++++++ ++++ l++lqD+tlgSLLs+lmqhcdppqr++plekg++pPWWPtG+e+ww +lgl+k+q ppykkp
AA31G00568 125 KFDKNGPAAIAKYEEECLAFGKSDGN----RMSQFVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGTPPPWWPTGNEEWWVKLGLPKSQT-PPYKKP 218
********************988877....7899*******************************************************998.9***** PP
GG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXX...XX...XXXX...XXXXXXXXXXXXXXXXXXX CS
EIN3 199 hdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsah...ss...slrk...qspkvtlsceqkedvegkk 288
hdlkk+wkv+vLtavi+hmsp+i++i++++rqsk+lqdkm+akes+++l+vlnqee+++++ s++ s+ ++r+ +++k ++ +++++dv+g +
AA31G00568 219 HDLKKMWKVGVLTAVINHMSPDIAKIKRHVRQSKCLQDKMTAKESAIWLAVLNQEESLIQQPSSDngtSNvteTHRRgnaDRRKPVVMSDSDYDVDGAE 317
*****************************************************************654227773333455555555666669****555 PP
XXX.X..XXXXXXXXX..................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXXXXXX CS
EIN3 289 eskik..hvqavktta..................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgsetelifadknsisqn 352
+ + + ++++++++ +++ +r++k+++ +s v + +++q++ ++ ++++d+n+++ +
AA31G00568 318 DASGSgsVSSKDNRRNqeppaatishsvrdkdkdKTEKHRRKKRPRIRSGNV------NQQEEEQLEIEQRDVVLPDMNHVDAH 395
5444411334444444899999999988876554555555555455555544......555899999999*********99988 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 4.6E-125 | 31 | 274 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 2.6E-71 | 150 | 282 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 9.94E-61 | 155 | 277 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 579 aa Download sequence Send to blast |
MDFDRMDNEP DDLASDNVAE IEVSDEEIDA EDLEKRMWKD RVRLKRIKER QKIGSQVTKE 60 TPKKISDQAQ RKKMSRAQDG ILKYMLKLME VCKVRGFVYG IIPEKGKPVS GSSDNIRAWW 120 KEKVKFDKNG PAAIAKYEEE CLAFGKSDGN RMSQFVLQDL QDATLGSLLS SLMQHCDPPQ 180 RKYPLEKGTP PPWWPTGNEE WWVKLGLPKS QTPPYKKPHD LKKMWKVGVL TAVINHMSPD 240 IAKIKRHVRQ SKCLQDKMTA KESAIWLAVL NQEESLIQQP SSDNGTSNVT ETHRRGNADR 300 RKPVVMSDSD YDVDGAEDAS GSGSVSSKDN RRNQEPPAAT ISHSVRDKDK DKTEKHRRKK 360 RPRIRSGNVN QQEEEQLEIE QRDVVLPDMN HVDAHMLEYN IGEGVVEPNI ALGLVENGLD 420 NHSQLVAPDF DNNNYSYLPS VDEMPAQTMI PVQERPMIYG PNANPELQYG SGYNFYNPSS 480 VFVHNQEEDI HTQLEMNVPA PNSSFEAPGG LQPVGLHGNE NGTSGDLPIL QNEQDKLLDT 540 NILSPFHDLP FDTGTFYSGF DTYGAFDDEF DKDFSWFGA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 3e-85 | 154 | 285 | 9 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 347 | 359 | KDKDKTEKHRRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
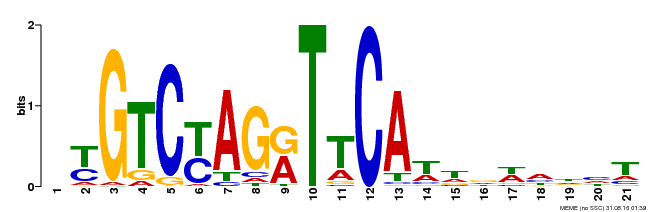
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA31G00568 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006390517.1 | 0.0 | ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 0.0 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | E4MXJ8 | 0.0 | E4MXJ8_EUTHA; mRNA, clone: RTFL01-23-B16 | ||||
| TrEMBL | V4KKK7 | 0.0 | V4KKK7_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006390517.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4658 | 25 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 0.0 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



