 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA32G00604 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 729aa MW: 83916.4 Da PI: 6.7443 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 31.5 | 3.2e-10 | 154 | 198 | 5 | 54 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
Er RR ++N++++ L+ l+P++ +K + a+iL ++YI++L
AA32G00604 154 PFTTERERRCHLNERYDALKLLIPNP-----TKGDRASILADGIDYINEL 198
5678**********************.....9***************998 PP
| |||||||
| 2 | HLH | 28.1 | 3.5e-09 | 532 | 572 | 9 | 54 |
HHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 9 ErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
Er RR + N +f L++l+P++ +K + a+++ A+eYIk+L
AA32G00604 532 ERERRVHFNARFIDLKKLIPSP-----TKSDRASTVGDAIEYIKEL 572
9*********************.....9****************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 9.2E-11 | 147 | 207 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 5.48E-10 | 147 | 202 | No hit | No description |
| SuperFamily | SSF47459 | 4.58E-13 | 148 | 219 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 13.821 | 149 | 198 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 6.1E-8 | 152 | 198 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.1E-8 | 155 | 204 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 12.697 | 523 | 572 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.7E-11 | 526 | 589 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.3E-5 | 529 | 578 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.6E-9 | 531 | 583 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 9.3E-7 | 531 | 572 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.76E-7 | 532 | 577 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0048658 | Biological Process | anther wall tapetum development | ||||
| GO:0052543 | Biological Process | callose deposition in cell wall | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 729 aa Download sequence Send to blast |
MEATIDNNGN SFNELTSTDL IHQMIQESSS FDQPNWVTES HDFTNSGFLM NPLQDHHHNS 60 NTNSSLPDLL NLLNLPKCSI SLPNSNLSDI ITTSCLTYDP LYHLNPPSQT PMIHMNPLET 120 QENENENENV IFEFSKDING CLQRKGRGKR KNKPFTTERE RRCHLNERYD ALKLLIPNPT 180 KGDRASILAD GIDYINELRR RVDELKYLVE KKRCGRHKNK RQKTQEDHKN SNNDDEEMEK 240 KVEIGLNDQC YSNKNLLRCS WLQRKSKVTE VDVRIVDDEV TIKIMQKKKI NCLFFVSKVI 300 DELQLDLHHV TGGQIGEHYS FLFNTKIYEG SIIYAKELSE FHLSPQDSAP VTASMQFQIN 360 PPCDQFHNNN INLMHQTVHD SSYTHPPQAN WDTNYQDFIN LGPNSTTPDL LSLLHLPRCS 420 LPPSMLPNSS ISFPNSTFSD IMSSSAVMYD PLFHLNFPLQ PRDQAFQHNQ RNGSSSLLGV 480 EDHQIEANRG ENPMFYQENN QMQFENGILD YNNGLKRKGR GSSGKTRIFP TERERRVHFN 540 ARFIDLKKLI PSPTKSDRAS TVGDAIEYIK ELLRSIEYLK MLLENKRLGK QRNKRARTAA 600 EEEDDTVNYR PQSEVDQTNF SKNNNSLRCS WLKRKSKVTE IDVRIIDDEV TIKLVQKKKI 660 NCLLFTTKVL DQLQLDLHHV AGGQIGEYYS FLFNTKISEG SCVYASGIAD TVMEVVEKQY 720 MEALPNNGY |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 198 | 222 | RRRVDELKYLVEKKRCGRHKNKRQK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00285 | DAP | Transfer from AT2G31220 | Download |
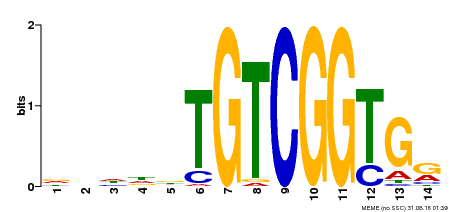
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA32G00604 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024003913.1 | 1e-175 | transcription factor bHLH10 | ||||
| Swissprot | Q84TK1 | 1e-169 | BH010_ARATH; Transcription factor bHLH10 | ||||
| TrEMBL | V4M9D8 | 1e-174 | V4M9D8_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006410254.1 | 1e-174 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1355 | 26 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G31220.1 | 1e-146 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



