 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA32G00958 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 351aa MW: 40089.2 Da PI: 5.7644 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 155.2 | 2.9e-48 | 14 | 140 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.kkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkge 98
l pGfrF+Ptd el+++yL++k++g+++++ vi++v+iyk+ePwdLp ++ ++e+ew++F+ r +ky++g++++rat+ gyWkatgk+++v+s +++
AA32G00958 14 LFPGFRFSPTDIELISYYLRRKIDGDENSV-AVIAQVEIYKFEPWDLPgESKLKSENEWFYFCARGRKYPHGSQSRRATQLGYWKATGKERNVKS-GNQ 110
579************************999.89***************543334678**************************************.999 PP
NAM 99 lvglkktLvfykgrapkgektdWvmheyrl 128
vg+k+tLvf+ grap+ge+t+W+mhey +
AA32G00958 111 IVGTKRTLVFHIGRAPRGERTEWIMHEYCI 140
****************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 2.62E-54 | 6 | 156 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 52.725 | 14 | 157 | IPR003441 | NAC domain |
| Pfam | PF02365 | 4.3E-24 | 16 | 139 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009845 | Biological Process | seed germination | ||||
| GO:0009938 | Biological Process | negative regulation of gibberellic acid mediated signaling pathway | ||||
| GO:0033619 | Biological Process | membrane protein proteolysis | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0071472 | Biological Process | cellular response to salt stress | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 351 aa Download sequence Send to blast |
MSKEAEMSIA VSTLFPGFRF SPTDIELISY YLRRKIDGDE NSVAVIAQVE IYKFEPWDLP 60 GESKLKSENE WFYFCARGRK YPHGSQSRRA TQLGYWKATG KERNVKSGNQ IVGTKRTLVF 120 HIGRAPRGER TEWIMHEYCI NDTPSQDALV VCRLRKNTDF RTTSTRKLVE DNVDVPEPDT 180 ARNSYSPYET EHKLLNEELA ESSNAFEEQV DDSDDDCYAE IMKDDIINLD EAAAAAWKTT 240 GEVLKANQTV LTNHGPISQE TLVRETAAAL TEKCDIKNES NQRMNCYALF KINNNEDLAA 300 GCRIFNHFRI KNYVDIRQKL KNVITNTVFL VTVLLSVFWV VLTAKEKGVV V |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 3e-42 | 13 | 157 | 19 | 168 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator activated by proteolytic cleavage through regulated intramembrane proteolysis (RIP), probably via metalloprotease activity. Regulates gibberellic acid-mediated salt-responsive repression of seed germination and flowering via FT, thus delaying seed germination under high salinity conditions. {ECO:0000269|PubMed:17410378, ECO:0000269|PubMed:18363782, ECO:0000269|PubMed:19704528, ECO:0000269|PubMed:19704545}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00280 | DAP | Transfer from AT2G27300 | Download |
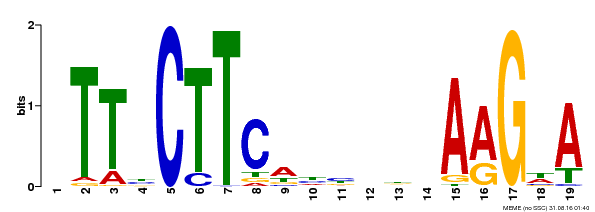
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA32G00958 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By high salt stress (PubMed:19704545, PubMed:17410378, PubMed:18363782, PubMed:19704528). Repressed by gibberellic acid (GA), but induced by the GA biosynthetic inhibitor paclabutrazol (PAC) (PubMed:18363782). Accumulates transiently in seeds upon imbibition (PubMed:19704545, PubMed:17410378, PubMed:18363782, PubMed:19704528). Induced by drought stress (PubMed:17158162). {ECO:0000269|PubMed:17158162, ECO:0000269|PubMed:17410378, ECO:0000269|PubMed:18363782, ECO:0000269|PubMed:19704528, ECO:0000269|PubMed:19704545}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_180298.1 | 1e-152 | NTM1-like 8 | ||||
| Swissprot | Q9XIN7 | 1e-153 | NAC40_ARATH; NAC domain-containing protein 40 | ||||
| TrEMBL | A0A1J3G9A5 | 1e-155 | A0A1J3G9A5_NOCCA; Protein NTM1-like 8 (Fragment) | ||||
| STRING | AT2G27300.1 | 1e-151 | (Arabidopsis thaliana) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10453 | 25 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G27300.1 | 1e-146 | NTM1-like 8 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



