 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA40G00266 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 386aa MW: 42611.8 Da PI: 6.894 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 56.4 | 7.1e-18 | 40 | 90 | 2 | 56 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkklege 56
y+GVr++ +g+WvAeIr+p + r +r +lg+f t +Aa a+++a+ kl+g+
AA40G00266 40 TYRGVRQRT-WGKWVAEIREP---N-RgTRLWLGTFNTSLQAALAYDQAAIKLYGH 90
69****999.**********8...3.35*************************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00018 | 1.26E-25 | 39 | 97 | No hit | No description |
| PROSITE profile | PS51032 | 21.746 | 40 | 97 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 8.4E-30 | 40 | 98 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 2.4E-34 | 40 | 103 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 2.16E-20 | 40 | 98 | IPR016177 | DNA-binding domain |
| Pfam | PF00847 | 2.4E-13 | 40 | 90 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-9 | 41 | 52 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-9 | 63 | 79 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 386 aa Download sequence Send to blast |
METQQQEQNN QVTPKKRGNM GRSRKGCMKG KGGPENATCT YRGVRQRTWG KWVAEIREPN 60 RGTRLWLGTF NTSLQAALAY DQAAIKLYGH QAKLNLPLQQ QQQFLHNSST LSFSSSSSSS 120 LSRVSSRDYQ SRCQSQSPCF SSSSNLCWLL PKQHQILQED QESVGFNETA TFRGKEKARF 180 GFNETATFRG KGGVGFNETK TFRGKEDGCN ETVTFRGKGG GVGFNETTTF RGKEDGFNET 240 VTFRGKEEEG VGFNETVTFS SKGEEGVNET VAFRGKGEGV NETVAFRGKE GGSWSRFLVG 300 EERKVEGGDV SMSSSSCGST ENKESLGVAI AVGNGGSGEG SSTGYLEMDD LLEIDDLGML 360 IEKNGDFKNW CSDDFQHPWN NNYNWY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gcc_A | 9e-18 | 41 | 99 | 3 | 62 | ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]CCGAC-3'. {ECO:0000250, ECO:0000269|PubMed:11798174}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00519 | DAP | Transfer from AT5G18450 | Download |
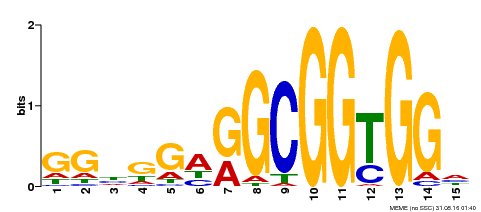
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA40G00266 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022563474.1 | 2e-54 | dehydration-responsive element-binding protein 2G-like | ||||
| Swissprot | P61827 | 3e-53 | DRE2G_ARATH; Dehydration-responsive element-binding protein 2G | ||||
| TrEMBL | A0A087G882 | 5e-57 | A0A087G882_ARAAL; Uncharacterized protein | ||||
| STRING | A0A087G882 | 8e-58 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9978 | 20 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18450.1 | 1e-47 | ERF family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



