 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA44G00405 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 933aa MW: 104228 Da PI: 5.1221 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.1 | 8.6e-18 | 36 | 82 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg WT+eEde l +av+ + g++Wk+Ia+++ Rt+ qc +rwqk+l
AA44G00405 36 RGQWTAEEDEVLRKAVHSFNGKNWKKIAEYFK-DRTDVQCLHRWQKVL 82
89*****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 59.6 | 6.7e-19 | 88 | 134 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd++++++v + G++ W+tIar+++ gR +kqc++rw+++l
AA44G00405 88 KGPWTKEEDDKIIELVGEIGPKKWSTIARYLP-GRIGKQCRERWHNHL 134
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 43.3 | 8.2e-14 | 140 | 182 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++++G++ W+ + ++ gR+++ +k++w+
AA44G00405 140 KEAWTQEEELVLIRAHQLYGNR-WAELTKFLP-GRSDNGIKNHWH 182
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.716 | 31 | 82 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.0E-15 | 33 | 89 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-14 | 35 | 84 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.5E-15 | 36 | 82 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.5E-22 | 37 | 90 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.29E-13 | 39 | 82 | No hit | No description |
| PROSITE profile | PS51294 | 31.661 | 83 | 138 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.49E-30 | 85 | 181 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.5E-17 | 87 | 136 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-17 | 88 | 134 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.45E-14 | 90 | 134 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-26 | 91 | 137 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-20 | 138 | 189 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.4E-12 | 139 | 187 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 18.314 | 139 | 189 | IPR017930 | Myb domain |
| Pfam | PF00249 | 4.8E-12 | 140 | 182 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.17E-9 | 142 | 182 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 933 aa Download sequence Send to blast |
MEGDCTPSLT PPEGLIDANQ RSRHGRTTGP ARRSTRGQWT AEEDEVLRKA VHSFNGKNWK 60 KIAEYFKDRT DVQCLHRWQK VLNPELVKGP WTKEEDDKII ELVGEIGPKK WSTIARYLPG 120 RIGKQCRERW HNHLNPAINK EAWTQEEELV LIRAHQLYGN RWAELTKFLP GRSDNGIKNH 180 WHSSVKKKLE SYVSSGLLDQ CQTLPLTPYE RSSMLPSAVM QRTIDENGCL SVNSENCQNS 240 SMVGSSLSAS DYPNQVINIG HHFQNSQENE QAASVTYQTD QYFYPDLEDI SISISDISYD 300 MEDCSQFTDH NVATSSSQDY QFDIQELSDI SHEMTQNTSD VPIPYTKDSK KESNNSANEM 360 IPDTECCRVL FQDQESEGIS ASKSLTQEPN EVDQADYQDP TLCASASDNQ RSESTKSPLQ 420 SCSSRSIATA ASGKETLRPA PLIISPDKHA NETSSRLIFH PSAATPKRRA NANGSFICTG 480 DPSSSTCGDQ DNNNSSQQDQ SYRVSDSKKL VAVNDFASLA EVKEHSRPDH QPNMTNQQHH 540 EDMDASSLCF PSLDLPVFSS DLLQSKNDLL QDYSPLGIRK LLMSTVTCMS PLRLWESPTG 600 KKTLVGKQSI LKKRTRDLLT PISEKRSDKK LEIDIAASLA NDFSRLEVMF DESCDRESIS 660 GEKTSVNLGE NHHQTFNEDD EESWRMKPSR KSLEKAEQTS MEEHAKGNDD SEPDVVHTES 720 LSGVLAEHNT NKLELTTPSK SVTKSVKGQG STPRNHLQRT LIATSNKEGL CLVLNSPSRT 780 RNTEEINHEN FSIFCGTPFR RGLESPSAWK SPFYMNSLLP SPRFNTDITI EDIGYIFSPG 840 ERSYNGGGPM TDNNNNEAAQ AAADALADIM EFSGSIILRE DKMKQKELNK ENDEPTMTER 900 RVLDFDCLSP TKPTNSSETS IAAVSSSSSS LKG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 4e-66 | 36 | 189 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 4e-66 | 36 | 189 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (PubMed:21862669). Involved in the regulation of cytokinesis, probably via the activation of several G2/M phase-specific genes transcription (e.g. KNOLLE) (PubMed:17287251, PubMed:21862669, PubMed:25806785). Required for the maintenance of diploidy (PubMed:21862669). {ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:21862669, ECO:0000269|PubMed:25806785}.; FUNCTION: Involved in transcription regulation during induced endoreduplication at the powdery mildew (e.g. G.orontii) infection site, thus promoting G.orontii growth and reproduction. {ECO:0000269|PubMed:20018666}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
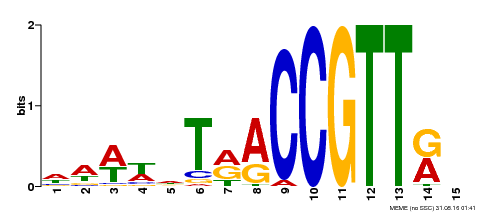
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA44G00405 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid (SA) (PubMed:16463103). Expressed in a cell cycle-dependent manner, with highest levels 2 hours before the peak of mitotic index in cells synchronized by aphidicolin. Activated by CYCB1 (PubMed:17287251). Accumulates at powdery mildew (e.g. G.orontii) infected cells. {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:17287251}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF371975 | 0.0 | AF371975.2 Arabidopsis thaliana putative c-myb-like transcription factor MYB3R-4 (MYB3R4) mRNA, complete cds. | |||
| GenBank | AY519650 | 0.0 | AY519650.1 Arabidopsis thaliana MYB transcription factor (At5g11510) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006399657.1 | 0.0 | transcription factor MYB3R-4 | ||||
| Swissprot | Q94FL9 | 0.0 | MB3R4_ARATH; Transcription factor MYB3R-4 | ||||
| TrEMBL | A0A087G6B1 | 0.0 | A0A087G6B1_ARAAL; Myb-related protein 3r-1-like | ||||
| STRING | A0A087G6B1 | 0.0 | (Arabis alpina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13213 | 18 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 0.0 | myb domain protein 3r-4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



