 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA4G00267 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 287aa MW: 32439.9 Da PI: 6.5889 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 102.8 | 2.2e-32 | 63 | 117 | 3 | 58 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRreve 58
++rY+eClkN+A+++Ggha+DGCgEfmp+ g+egt+++lkCaAC+CHRnFHR+e++
AA4G00267 63 RFRYRECLKNQAVNIGGHALDGCGEFMPA-GSEGTIESLKCAACNCHRNFHRKETN 117
789*************************9.999********************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04770 | 6.4E-29 | 64 | 116 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 9.4E-30 | 65 | 115 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| ProDom | PD125774 | 4.0E-22 | 65 | 115 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 26.294 | 66 | 115 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-28 | 172 | 245 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.79E-17 | 172 | 245 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 4.6E-26 | 185 | 240 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010371 | Biological Process | regulation of gibberellin biosynthetic process | ||||
| GO:0010431 | Biological Process | seed maturation | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MEFNDDEEDE AQEEEEEMNL PQNDTVFTSP HLETTTTTTT STTSGGLMVL HTGTPTGNTT 60 GTRFRYRECL KNQAVNIGGH ALDGCGEFMP AGSEGTIESL KCAACNCHRN FHRKETNHPI 120 PPPGFYRIPT PVTTYHQRPI LPPPAPPLAL PPPPIRERSD LEDELNNKMD TSSAEGGGGN 180 MMRNKRFRTK FTAEQKEMML GLAERIGWRI QRQDDELISR FCQESGVSRQ VLKVWLHNNK 240 HTLGKSSSST TSPPPINISH HHHQHQQHQH HHHQQNQSLF HQEQDQP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wh5_A | 3e-29 | 185 | 242 | 18 | 75 | ZF-HD homeobox family protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00588 | DAP | Transfer from AT5G65410 | Download |
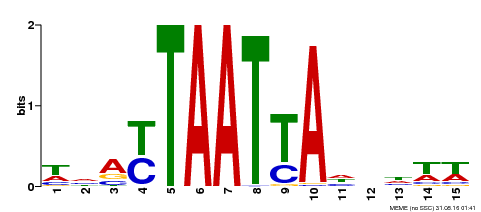
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA4G00267 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018437622.1 | 3e-92 | PREDICTED: zinc-finger homeodomain protein 1-like | ||||
| Swissprot | Q9FKP8 | 4e-83 | ZHD1_ARATH; Zinc-finger homeodomain protein 1 | ||||
| TrEMBL | A0A398AN32 | 3e-91 | A0A398AN32_BRACM; Uncharacterized protein | ||||
| STRING | Bra031859.1-P | 3e-89 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13237 | 17 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65410.1 | 8e-77 | homeobox protein 25 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



