 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA52G00002 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 253aa MW: 28727.2 Da PI: 6.5178 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 29.1 | 2.4e-09 | 24 | 71 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT....-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGgg....tWktIartmgkgRtlkqcksrwqky 47
W++eE + + +a + + + +W +Ia++++ g++ ++ + ++k+
AA52G00002 24 QWSKEENKMFEKALAIYTEEdspdRWFKIAEMIP-GKSVVDVMNQYNKL 71
7*****************99**************.************97 PP
| |||||||
| 2 | Myb_DNA-binding | 40 | 9.2e-13 | 125 | 169 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + ++G+g+W+ I+r + +t+ q+ s+ qky
AA52G00002 125 PWTEEEHRRFLLGLLKYGKGDWRNISRNFVVSKTPTQVASHAQKY 169
8*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 9.293 | 20 | 74 | IPR017884 | SANT domain |
| SMART | SM00717 | 6.7E-9 | 21 | 74 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 9.33E-11 | 23 | 80 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 9.0E-5 | 24 | 71 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.9E-8 | 24 | 71 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.27E-9 | 25 | 71 | No hit | No description |
| PROSITE profile | PS51294 | 18.771 | 118 | 174 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.37E-15 | 120 | 173 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.1E-11 | 122 | 173 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.1E-17 | 122 | 173 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 1.7E-10 | 122 | 172 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.12E-9 | 125 | 169 | No hit | No description |
| Pfam | PF00249 | 4.0E-10 | 125 | 169 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 253 aa Download sequence Send to blast |
METLQPLSHH FINEDLMNLQ SSSQWSKEEN KMFEKALAIY TEEDSPDRWF KIAEMIPGKS 60 VVDVMNQYNK LEEDVLDIES GRVPIPGYPP ACSSDPLGFE RDVACRKRSN GNSKGCDQER 120 RKGVPWTEEE HRRFLLGLLK YGKGDWRNIS RNFVVSKTPT QVASHAQKYY QRQISGTKDK 180 RRPSIHDITT VNLLNANLMR SSSEFNPDLG FNSDEGMMFS RQNMLSESLF SPSISHFGDA 240 GKFTGENCFG AGG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 2e-14 | 21 | 88 | 7 | 73 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
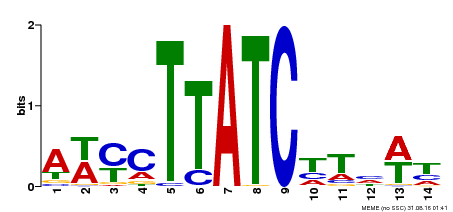
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00347 | DAP | Transfer from AT3G11280 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA52G00002 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010486776.1 | 1e-122 | PREDICTED: transcription factor DIVARICATA | ||||
| Swissprot | Q8S9H7 | 3e-70 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | A0A3N6Q551 | 1e-119 | A0A3N6Q551_BRACR; Uncharacterized protein | ||||
| TrEMBL | V4M940 | 1e-119 | V4M940_EUTSA; Uncharacterized protein | ||||
| STRING | XP_010486776.1 | 1e-122 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4340 | 26 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G11280.2 | 1e-123 | MYB family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



