 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA53G00325 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 470aa MW: 52500.3 Da PI: 6.7418 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.9 | 1.6e-12 | 323 | 369 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI ++q
AA53G00325 323 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLADAITYITDMQ 369
799***********************66......***************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 6.3E-41 | 48 | 227 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.234 | 319 | 368 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.96E-17 | 319 | 385 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.60E-14 | 322 | 373 | No hit | No description |
| Pfam | PF00010 | 4.2E-10 | 323 | 369 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.2E-16 | 323 | 384 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.6E-14 | 325 | 374 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 470 aa Download sequence Send to blast |
MGLKFWENQD DRVMVESTIG SEACDFFISS ASNSVLTKLV PPLSDPNLQQ GLRHLVEGSE 60 WDYAIFWLAS NVNSSDGCVL IWGDGHCRLP KVKKGISGED SSKLDETKRR VLQKLHLSFV 120 GSDEEHRLPK SGPLSDLEMF HLASMYFSFR CDTSKYGPAG TYVSGKPLWA ADLPSCLSYY 180 RVRSFLGRSA GFQTVLSVPV NSGVVELGSL RQIPEDKSVI EMVKAVFGGS DFVQAKEAPK 240 IFGRQLSLGN GGSKPRSMSI NFSPKMEDDT GFSLESYDVQ AIGGSNQVYG YEQGKDEALY 300 LTDEQKPRKR GRKPANGREE ALNHVEAERQ RREKLNQRFY ALRAVVPNIS KMDKASLLAD 360 AITYITDMQK KIRVYETEKQ IMSRRESNQI TPADIDFQQR HEDGIVRLSC PLETHPVSSV 420 IQTFREHELT PHDANVAVTE DGVVHTFTVR PQAGFTAEQL KDKLVASFAQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_B | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_E | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_F | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_G | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_I | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_M | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| 5gnj_N | 4e-26 | 317 | 402 | 2 | 83 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
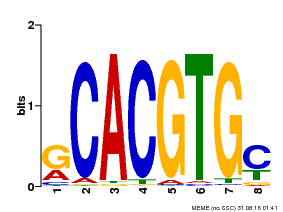
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA53G00325 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_002870159.1 | 0.0 | transcription factor bHLH3 | ||||
| Swissprot | O23487 | 0.0 | BH003_ARATH; Transcription factor bHLH3 | ||||
| TrEMBL | D7M9V3 | 0.0 | D7M9V3_ARALL; Basic helix-loop-helix family protein | ||||
| STRING | scaffold_702851.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6611 | 27 | 41 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 0.0 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



