 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA53G00686 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 579aa MW: 62740.8 Da PI: 9.4319 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 16.2 | 3e-05 | 77 | 99 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+ C+ C+k F r nL+ H r H
AA53G00686 77 FICEVCNKGFQREQNLQLHRRGH 99
89*******************88 PP
| |||||||
| 2 | zf-C2H2 | 13.1 | 0.00028 | 153 | 175 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
+kC++C+k++ +s+ k H +t+
AA53G00686 153 WKCEKCSKRYAVQSDWKAHSKTC 175
58*****************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:3.30.160.60 | 6.5E-6 | 76 | 99 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.15E-7 | 76 | 99 | No hit | No description |
| SMART | SM00355 | 0.0052 | 77 | 99 | IPR015880 | Zinc finger, C2H2-like |
| Pfam | PF12171 | 2.8E-5 | 77 | 99 | IPR022755 | Zinc finger, double-stranded RNA binding |
| PROSITE profile | PS50157 | 10.99 | 77 | 99 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 79 | 99 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 110 | 118 | 148 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.6E-5 | 141 | 174 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 3.15E-7 | 148 | 173 | No hit | No description |
| SMART | SM00355 | 130 | 153 | 173 | IPR015880 | Zinc finger, C2H2-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 579 aa Download sequence Send to blast |
MAASSSSAAS FFGVRQDDQS HHLPLPPSSS AAGAAAPPPQ LLETQPPKKK RNQPRTPNSD 60 AEVIALSPKT LMATNRFICE VCNKGFQREQ NLQLHRRGHN LPWKLKQKST KEVKRKVYLC 120 PEPTCVHHDP SRALGDLTGI KKHYYRKHGE KKWKCEKCSK RYAVQSDWKA HSKTCGTKEY 180 RCDCGTLFSR RDSFITHRAF CDALAQESAR HPTTTLTSLP SHHFPYGQSN TNNTNNNTNT 240 SSMLLGLSHL DHHHHHQTGD VLRLGNTGGG SVSRASTDLI NASNYFMQEQ QQQNPSFQDH 300 QQGYLGNNKN QNSAMSYHNL MQFSHDHQNH HNNSQSSNLF NLSFLNNGAP TATSNVAVSS 360 GNNLMISNPF DGENAVNLGG GDQGTTSTGL FPNNLMNSGD RINSGAVPSL FSSSLQNTNS 420 SPHMSATALL QKAAQMSSTS SGTNNNNNNN NNNASSILRS FGSGIYGENE SNLHDLMNSF 480 STPGPGNGVD STFGSYGGIN KGLNVDNKSQ NMTRDFLGVG QIVRSMSGGG ATGGFQQQQQ 540 HGNSRERLGS SSDSADRNSL PGGAPASSPP YGSMHHGSF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5b3h_C | 1e-33 | 149 | 210 | 3 | 64 | Zinc finger protein JACKDAW |
| 5b3h_F | 1e-33 | 149 | 210 | 3 | 64 | Zinc finger protein JACKDAW |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor acting as a positive regulator of the starch synthase SS4. Controls chloroplast development and starch granule formation (PubMed:22898356). Binds DNA via its zinc fingers (PubMed:24821766). Recognizes and binds to SCL3 promoter sequence 5'-AGACAA-3' to promotes its expression when in complex with RGA (PubMed:24821766). {ECO:0000269|PubMed:22898356, ECO:0000269|PubMed:24821766}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00255 | DAP | Transfer from AT2G02070 | Download |
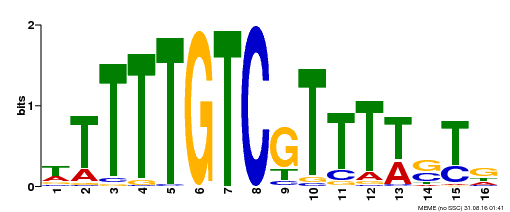
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA53G00686 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_018434643.1 | 0.0 | PREDICTED: protein indeterminate-domain 5, chloroplastic-like | ||||
| Swissprot | Q9ZUL3 | 0.0 | IDD5_ARATH; Protein indeterminate-domain 5, chloroplastic | ||||
| TrEMBL | M4ECV7 | 0.0 | M4ECV7_BRARP; Uncharacterized protein | ||||
| STRING | Bra026617.1-P | 0.0 | (Brassica rapa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4373 | 26 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02070.1 | 0.0 | indeterminate(ID)-domain 5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



