 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA53G01286 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 390aa MW: 41930.9 Da PI: 7.1324 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.9 | 5.2e-39 | 57 | 118 | 2 | 63 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkksss 63
ke+al+cprC+s +tkfCyynnysl+qPryfCk CrryWt+GG+lrn+P+Ggg+rknk+sss
AA53G01286 57 KEQALNCPRCNSIHTKFCYYNNYSLTQPRYFCKDCRRYWTEGGSLRNIPIGGGSRKNKRSSS 118
7899******************************************************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 5.0E-29 | 50 | 115 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 7.3E-33 | 59 | 115 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.602 | 61 | 115 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 63 | 99 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 390 aa Download sequence Send to blast |
MDTAKWPQEF VVKPMNEIVT NTCLKQSLPL PPPPPPPQTV GAVVGSTVER KMTRPQKEQA 60 LNCPRCNSIH TKFCYYNNYS LTQPRYFCKD CRRYWTEGGS LRNIPIGGGS RKNKRSSSSS 120 TTTTTTSSSS SNSNSSPSKK QLFATQNPNL NEGKVQVQDL TFSQGLNTNP HEVKDLNLAF 180 SQGFGIAEFL QVPSIKNNNN SNPLVSTSSA LELLGISNSS GSSSIQRPSF IGFPHVNDPS 240 SVYTPSGSGF GLSYPPFQEF MRPPTLGFSL DTNGGYNGEP LNQQEGSNSN INNGNNTNGI 300 PLLPFESLKL PVSSSSTNSS GGGNDHHHHQ QHQHDNHQEQ QHNQGNSDHE EEKEEGGDQS 360 VGFWSGMLGV GVGGGGAGAT TASGGGTWQL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
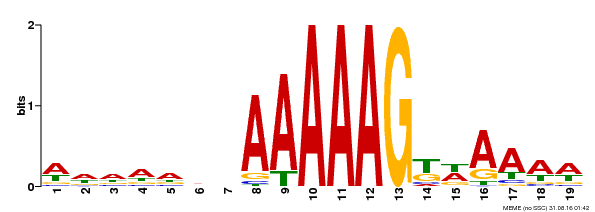
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00210 | DAP | Transfer from AT1G64620 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA53G01286 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006391598.1 | 1e-122 | dof zinc finger protein DOF1.8 | ||||
| Swissprot | Q84JQ8 | 1e-112 | DOF18_ARATH; Dof zinc finger protein DOF1.8 | ||||
| TrEMBL | V4KCL5 | 1e-121 | V4KCL5_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006391598.1 | 1e-122 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14932 | 17 | 20 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G64620.1 | 1e-100 | Dof family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



