 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA56G00010 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 610aa MW: 66615.2 Da PI: 4.7554 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.1 | 1.3e-12 | 435 | 480 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
AA56G00010 435 NHVEAERQRREKLNQRFYSLRAVVPNV-----SKMDKASLLGDAIAYINEL 480
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 9.6E-54 | 50 | 236 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.003 | 431 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.53E-18 | 434 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 1.91E-14 | 434 | 485 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 5.0E-18 | 435 | 501 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.3E-10 | 435 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.2E-16 | 437 | 486 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 610 aa Download sequence Send to blast |
MLRISNQITN AILFSVSSSL FTRHVITTTS LSTFSPESLH LLSESLDDTL QQRLQALIEA 60 AGESWTYAIF WQISYDFDST GGDGENAAIL GWGDGFYKGE EDKEKKKPNP VSAAEQEHRK 120 RVIRELNSLI SGGNGVSDEA NDEDVTDTEW FFLVSMTQSF MNGVGLPGEA FLNSRLIWLS 180 GSGALVGSGC ERAGQGQIYG LQTMVCIPAE NGVVELGSSE VINQSSDLMD KVNSLFNFNN 240 GGGGEACSWG VNLNPDQGEN DPGMWISEPT PTGIVTPAIN TGNSIPNSDS HQISKLENAS 300 SVENPNRNQN SSLIERDLNF SSSGFDSSRP KSSEILSFCG NTNGDGIGFS GQTRFVADES 360 NKKRSPISKE SGNNNDDGML SFTCGTVIPS TAAKSGDSDH SDLEASVVKI PVVEPEKKPR 420 KRGRKPANGR EEPLNHVEAE RQRREKLNQR FYSLRAVVPN VSKMDKASLL GDAIAYINEL 480 KSKLEQTESE KEEIQKQLDG LRKEGNGKGC CAKDRKCQNS QDSSTISIEM EIDVKIIGWD 540 VMIRIQCSKK NHPGARFMDA LKELDMEVNH ASLSVVNDLM IQQATVKMGS QFFTHDQLKV 600 ALMTKVGENP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 1e-103 | 47 | 239 | 6 | 195 | Transcription factor MYC3 |
| 4rqw_B | 1e-103 | 47 | 239 | 6 | 195 | Transcription factor MYC3 |
| 4rs9_A | 1e-103 | 47 | 239 | 6 | 195 | Transcription factor MYC3 |
| 4yz6_A | 1e-103 | 47 | 239 | 6 | 195 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 416 | 424 | KKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in tryptophan, jasmonic acid (JA) and other stress-responsive gene regulation. With MYC2 and MYC4, controls additively subsets of JA-dependent responses. Can form complexes with all known glucosinolate-related MYBs to regulate glucosinolate biosynthesis. Binds to the G-box (5'-CACGTG-3') of promoters. Activates multiple TIFY/JAZ promoters. {ECO:0000269|PubMed:12136026, ECO:0000269|PubMed:21242320, ECO:0000269|PubMed:21321051, ECO:0000269|PubMed:21335373, ECO:0000269|PubMed:23943862}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00085 | PBM | Transfer from AT5G46760 | Download |
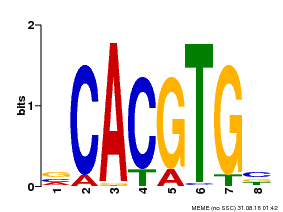
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA56G00010 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Barely up-regulated by jasmonic acid. {ECO:0000269|PubMed:21242320, ECO:0000269|PubMed:21335373}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_024010949.1 | 0.0 | transcription factor MYC3 | ||||
| Swissprot | Q9FIP9 | 0.0 | MYC3_ARATH; Transcription factor MYC3 | ||||
| TrEMBL | E4MW10 | 0.0 | E4MW10_EUTHA; mRNA, clone: RTFL01-05-E20 | ||||
| TrEMBL | V4L837 | 0.0 | V4L837_EUTSA; Uncharacterized protein | ||||
| STRING | XP_006398371.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1265 | 28 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G46760.1 | 0.0 | bHLH family protein | ||||



