 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA61G00372 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 936aa MW: 106167 Da PI: 5.94 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 134.2 | 4.1e-42 | 124 | 200 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cqve+C adls++k+yhrrhkvCe+hska+++lv g++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk +
AA61G00372 124 ICQVENCGADLSKVKDYHRRHKVCEMHSKATSALVGGVMQRFCQQCSRFHGLQEFDEGKRSCRRRLAGHNKRRRKGN 200
5*************************************************************************976 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51141 | 32.325 | 122 | 199 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:4.10.1100.10 | 3.2E-33 | 123 | 186 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 4.71E-39 | 124 | 203 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 5.0E-31 | 125 | 198 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 3.3E-8 | 716 | 831 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.81E-9 | 731 | 832 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 10.524 | 738 | 792 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 8.23E-9 | 738 | 828 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 936 aa Download sequence Send to blast |
MEAHYELSRM DLKGLGKRSV EWDLNDWKWD GDLFIGRKLN HGVNETMSRQ FFPLGSNSSS 60 SCSEEVNVEV LEKKRRVIVL DEENLRENGE DDARRLALKL GGNGETMENC ERNGVKKSKI 120 GGAICQVENC GADLSKVKDY HRRHKVCEMH SKATSALVGG VMQRFCQQCS RFHGLQEFDE 180 GKRSCRRRLA GHNKRRRKGN PETVSNVNPL SNEQTSNYLL ITLLKILSNL HCMLSFNFME 240 ISISIRRFYR FLILFYVAAN QSDSIVDQDL LSHLLKSIVN QTGDQIGKSL VELLQGQASL 300 NNGNSGLVLV EETPREDLKH TSVNVPETTW QEVRIAEDKS EKRVKMNDFD LNDIYIDSDE 360 TTDVERSPLA TTNPATSSLD YHQDSHQFSP PQTSRNSDSA SDQSPSSSMG DAQSRTDRIV 420 FKLFGKEPND FPVVLRGQIL DWLAHSPTDM ESYIRPGCIV LTIYLRQTEA AWEETHEKSL 480 FRLLFQLLCD LGFGLKQLLD ISDDPLWTTG WIYLRVQNQL AFAFNGEVVL DTSLPLRNQD 540 YSEIISVRPL AVSAKERVEF TVKGINLRRP GTRLLCAVEG TYLNQEATDE LISKNDTLEE 600 KNEIEIVKFS CEIPIENGRG FMEIEDQSLS SSFFPFIVSE DKEVCSEIRL LETTLEFTGT 660 DSAKQAMDFI HELGWLLHRT QLKNTSNPNS DQFPLQRFKW LIEFSMDREW CAVFKKLLNL 720 LLEETPSPEA TVSELCLLHR AVRRNSKSLV QMLLRFKNSL FRPDAAGPAG LTPLHIAAGK 780 DGSEDVLDAL TEDPGLVGIE AWKSSKDSTG FTPEDYARLR GHFSYIHLVQ RKIKKKNVVE 840 DHVVVDVLED LGDLKVEKRS SAMEITTQSV LSNCKVCDRK MVYGRARRSV VYRPAMLSMV 900 AIAAVCVCVA LLFKSCPEVL YVFQPFRWEL LAYGTS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-32 | 121 | 198 | 7 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00060 | PBM | Transfer from AT3G60030 | Download |
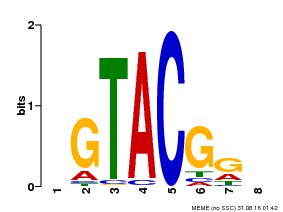
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA61G00372 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_191562.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Refseq | XP_002878311.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Refseq | XP_020880464.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Refseq | XP_020880465.1 | 0.0 | squamosa promoter-binding-like protein 12 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | A0A1J3IQZ6 | 0.0 | A0A1J3IQZ6_NOCCA; Squamosa promoter-binding-like protein 1 | ||||
| STRING | AT3G60030.1 | 0.0 | (Arabidopsis thaliana) | ||||
| STRING | scaffold_503227.1 | 0.0 | (Arabidopsis lyrata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



