 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA6G00064 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 676aa MW: 75753.7 Da PI: 5.5173 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 152.7 | 2.8e-47 | 82 | 228 | 1 | 145 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqsslks 100
gg+g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+V++AL+reAGw+v++DG+tyr++ p+ +++ +s+es+l+ss+
AA6G00064 82 GGKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVIAALAREAGWTVDADGNTYRQSHLPN-----HVQFPARSVESPLSSSTLK 176
6899********************************************************************9999.....6778888888888855555 PP
DUF822 101 salaspvesysaspksssfpspssldsislasa.......asllpvlsvlsl 145
+ ++ ++ +++ ++ ++ sp+slds+ +a++ + +p++sv ++
AA6G00064 177 NCAKAALDCQHSILRIDENFSPVSLDSVVIAENdpgsaiyTGASPITSVGCM 228
556778999*********************9998887777677777777776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.2E-44 | 83 | 213 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 1.65E-149 | 246 | 672 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 3.3E-156 | 251 | 673 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 1.4E-78 | 256 | 638 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 286 | 300 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 307 | 325 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 414 | 436 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 487 | 506 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 521 | 537 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 538 | 549 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 556 | 579 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 9.0E-48 | 595 | 617 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 676 aa Download sequence Send to blast |
MNTLNNNTTT MGPQDTIDPN SNPNPDPDQS IHQNQHHQPQ SRRPRGFAAA AASIGPAESD 60 VNNENSAGIG GGETSSGGGG GGGKGKRERE KEKERTKLRE RHRRAITSRM LAGLRQYGNF 120 PLPARADMND VIAALAREAG WTVDADGNTY RQSHLPNHVQ FPARSVESPL SSSTLKNCAK 180 AALDCQHSIL RIDENFSPVS LDSVVIAEND PGSAIYTGAS PITSVGCMET NQLIQDVHST 240 EPRNDFTGSF YVPVYAMLPV GIINNFGQLV DAEGVRQEIS YMKSLNVDGV VIDCWWGIVE 300 GWNPQKYVWS GYRELFNLIR DFKLKLQVVM AFHEYYGGNA SGNVMVPLPQ WVVEIGKDNP 360 DIYFTDREGR RNFECLNWSI DKERVYFDFM RSFRSEFDDL FEEGLIAAVE IGLGASGELK 420 YPSFPERLGW IYPGIGEFQC YDKYSQQNLL KEAKSRGFAI CGKGPENAGQ YNSRPHETGF 480 FQERGEYDSY YGRFFLNWYS QLLIGHADNV LSLATLAFED TKIIVKIPAI YWSYKTASHA 540 AELTAGYYNP SNRDGYTPVF ETLKKHSVTV KFICPGPQMP LPNEHEEALA DPEGLTWQVI 600 NAAWDKELLV GGENAVACFD REGSMRLVEL AKPRNHPDSY HFSFFTYRQP SPLLQGSTCF 660 PDLDYFIKRL HGEIQR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wqs_A | 1e-111 | 251 | 674 | 12 | 447 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
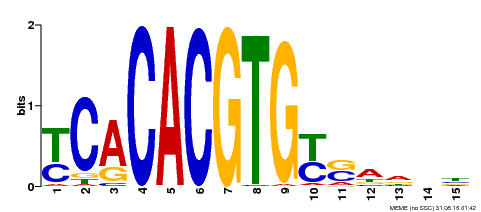
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA6G00064 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010494622.1 | 0.0 | PREDICTED: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | D7MUR1 | 0.0 | D7MUR1_ARALL; Beta-amylase | ||||
| STRING | XP_010494622.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



