 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA87G00136 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 205aa MW: 24029.3 Da PI: 7.1005 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.6 | 2.9e-18 | 15 | 62 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT eEd++l d+v+ +G g+W++Ia++ g++R++k+c++rw +yl
AA87G00136 15 KGLWTMEEDKILMDYVQSHGQGHWNRIAKKTGLKRCGKSCRLRWMNYL 62
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.3 | 6.1e-17 | 68 | 113 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T E++l+++++k+lG++ W++Ia++++ Rt++q+k++w+++l
AA87G00136 68 RGNFTDLEEDLIIRLHKLLGNR-WSLIAKRIP-ERTDNQVKNYWNTHL 113
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-24 | 8 | 65 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.681 | 10 | 62 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.95E-30 | 13 | 109 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-13 | 14 | 64 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-16 | 15 | 62 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.91E-10 | 18 | 62 | No hit | No description |
| PROSITE profile | PS51294 | 24.675 | 63 | 117 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-25 | 66 | 117 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-16 | 67 | 115 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.6E-16 | 68 | 113 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.97E-11 | 70 | 113 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010053 | Biological Process | root epidermal cell differentiation | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0048629 | Biological Process | trichome patterning | ||||
| GO:0090377 | Biological Process | seed trichome initiation | ||||
| GO:0090378 | Biological Process | seed trichome elongation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 205 aa Download sequence Send to blast |
MRKIIREVKD EEYKKGLWTM EEDKILMDYV QSHGQGHWNR IAKKTGLKRC GKSCRLRWMN 60 YLSPNVNRGN FTDLEEDLII RLHKLLGNRW SLIAKRIPER TDNQVKNYWN THLSKKLGLE 120 EHSSVTKPVC DDESTPSILT CTTHQDEKVT ITSFNSVADE FRLNLEPKPD QTTTIPNRFE 180 TFWVLEDEFE LSSLTMMDFT SGSCL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 4e-27 | 6 | 117 | 18 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator, when associated with BHLH2/EGL3/MYC146 or BHLH12/MYC1. Regulates the epidermal cell fate specification. Mediates the formation of columellae and accumulation of mucilages on seed coats. Controls the elongation of epidermal cells positively in roots but negatively in stems, leading to the promotion of primary roots elongation and repression of leaves and stems elongation, respectively. Ovoids ectopic root-hair formation, probably by inducing GL2 in roots. Controls trichome initiation and branching. {ECO:0000269|PubMed:11437443, ECO:0000269|PubMed:15361138, ECO:0000269|PubMed:15668208, ECO:0000269|PubMed:15728674}. | |||||
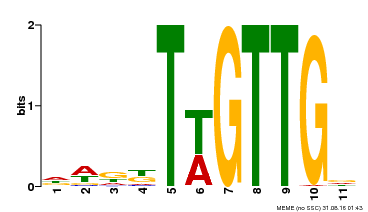
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00538 | DAP | Transfer from AT5G40330 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA87G00136 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010450681.1 | 1e-109 | PREDICTED: transcription factor MYB23 | ||||
| Swissprot | Q96276 | 1e-103 | MYB23_ARATH; Transcription factor MYB23 | ||||
| TrEMBL | A0A1J3HA71 | 1e-106 | A0A1J3HA71_NOCCA; Transcription factor MYB23 (Fragment) | ||||
| STRING | XP_010450681.1 | 1e-108 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4 | 28 | 2646 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G40330.1 | 1e-105 | myb domain protein 23 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



