 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AA96G00099 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Aethionemeae; Aethionema
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1327aa MW: 149880 Da PI: 7.458 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 14.6 | 9.4e-05 | 1210 | 1235 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y+C Cg+sF++k +L H ++ +
AA96G00099 1210 YVCDmeGCGMSFRSKKELALHKKNiC 1235
99*******************99866 PP
| |||||||
| 2 | zf-C2H2 | 14.7 | 9e-05 | 1235 | 1257 | 3 | 23 |
ET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 Cp..dCgksFsrksnLkrHirtH 23
Cp dCgk F ++ +L++H r+H
AA96G00099 1235 CPvkDCGKKFFSHKYLVQHRRVH 1257
9999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 12.7 | 0.00039 | 1293 | 1319 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
AA96G00099 1293 YVCGepGCGQTFRFVSDFSRHKRKtgH 1319
89********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 8.1E-17 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.728 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 8.1E-15 | 21 | 54 | IPR003349 | JmjN domain |
| SuperFamily | SSF51197 | 3.84E-28 | 115 | 177 | No hit | No description |
| SMART | SM00558 | 4.0E-52 | 197 | 366 | IPR003347 | JmjC domain |
| PROSITE profile | PS51184 | 34.029 | 200 | 366 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 3.84E-28 | 215 | 385 | No hit | No description |
| Pfam | PF02373 | 7.7E-38 | 230 | 349 | IPR003347 | JmjC domain |
| SMART | SM00355 | 6.3 | 1210 | 1232 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 0.0084 | 1233 | 1257 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.736 | 1233 | 1262 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 5.1E-5 | 1234 | 1256 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 1235 | 1257 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.98E-9 | 1249 | 1291 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 9.4E-6 | 1257 | 1270 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.468 | 1263 | 1292 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0011 | 1263 | 1287 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1265 | 1287 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 1.3E-4 | 1271 | 1285 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 2.43E-8 | 1281 | 1315 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.4E-9 | 1286 | 1316 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.54 | 1293 | 1319 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.907 | 1293 | 1324 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1295 | 1319 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0040010 | Biological Process | positive regulation of growth rate | ||||
| GO:0045815 | Biological Process | positive regulation of gene expression, epigenetic | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0071558 | Molecular Function | histone demethylase activity (H3-K27 specific) | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1327 aa Download sequence Send to blast |
MAVSEQSQDV FPWLKSLPLA PEFRPTLSEF QDPIAYIFKI EEEASKYGIC KIVPPVAVSS 60 KKSAISNLNR SLVARAASRV CKGGACDYDN GPTFTTRQQQ IGFCPRKQRP VQRPVWQSGE 120 HYSFGEFEAK AKNFERNYLK KSGKKSQLSP LEIETLYWRA TVDKPFSVEY ANDMPGSAFI 180 PLSTAAARRR ESGDGGTVGE TNWNMRAVAR ADGSLLKFMK EEIPGVTSPM VYIAMMFSWF 240 AWHVEDHDLH SLNYLHMGAG KTWYGVPKDA AAAFEEVVRV HGYGEELNPL VTFSTLGEKT 300 TVMSPEVFVK AGIPCCRLVQ NPGEFVVTFP RAYHSGFSHG FNCGEASNIA TPEWLKMAKD 360 AAIRRAAIKY PPMVSHLQLL YDFALALGSK VSTSMNTKPR SSRLKDKKRS EGERLTKELF 420 VRNVIHNNQL LHSLGKGSPV ALLPLSSSDI SVCSELRIGA QLGTNQEKIG EFNKDVMKSE 480 DLSSDMIGLS NGYTDSVSVK EKFTSLCEKS RSYKTEENLD RGVGLSDQRL FSCVTCGILS 540 YDCVAIVQPK EGAARYLMSA DCSFFNDWSG SSNLIQDSEG KEKRDNESLY NVKIGDERVA 600 TTFLTNSRKE DGALGLLASA YGDSSDSEDE NKALSVDVCA VSCDIELKQA RDCGNSDGFV 660 EVSHGSSSYS TISCKNTSVL EIGLPFVSRS DDDSSRLHVF CLEHAAEVEQ QLRPIGGIHL 720 TLLCHPEYPR IEAEAKSVAE ELGISHEWND TEFRNVNRED EERIQKALNN VEAKTGNSDW 780 AVKLGINLSY SAVLSRSPLY SKQMPYNSVI YNAFGRNSST NSPANPEGSS RRSSRQRKFV 840 VGKWCGKVWM SHQVHPFMLE EDLEEEEPEK PEKSLHLRAV LDEEDTEKKR PCNIFRNSVT 900 MVAKKYSRKR KMRAKTSTRK KVLKIENGVS DDTSEDHSYK QQWRASGNEE EDVYFETGNT 960 VSGDSYDQQL KNFRRNKRSS CRDQEIESDD EVSDRSLGEE YAVRECAASE SSMENSFQLY 1020 KEKQSINNKK AFGNSIYDQD QEDETLYRSQ SKGIPRSKRT KVFRNPISYD SEENGSFTRS 1080 IKQIRQAGNE YDSADEEDDR EGRDSPQDRD FIGTTKRVCK SRQTRANRKT KAETIQSLEE 1140 IKGRSFQKNQ ETDSCIDGPS TRLRMRKQKP LRGLEIKPRR IAVSEEEVEE EEEEVEEEED 1200 DIEEGESSGY VCDMEGCGMS FRSKKELALH KKNICPVKDC GKKFFSHKYL VQHRRVHLDD 1260 RPLKCPWKGC KRTFKWAWAR TEHIRVHTGA RPYVCGEPGC GQTFRFVSDF SRHKRKTGHS 1320 VKKTKKK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 3e-75 | 9 | 432 | 4 | 387 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 3e-75 | 9 | 432 | 4 | 387 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 906 | 911 | SRKRKM |
| 2 | 908 | 920 | KRKMRAKTSTRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
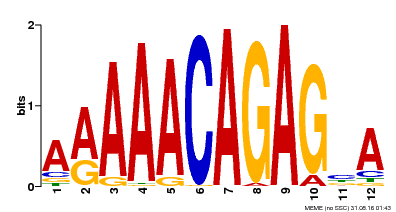
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | AA96G00099 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006404260.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Refseq | XP_024013589.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A1J3JIY4 | 0.0 | A0A1J3JIY4_NOCCA; Lysine-specific demethylase REF6 | ||||
| STRING | XP_006404260.1 | 0.0 | (Eutrema salsugineum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5259 | 28 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



