 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_000118-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 429aa MW: 47612.5 Da PI: 6.4081 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.7 | 9.4e-32 | 51 | 106 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQ.kYRl 55
kprl+Wt++LHerFveav+qLGG++kAtPkt+++lm+++gLtl+h+kSHLQ kYRl
AHYPO_000118-RA 51 KPRLKWTHDLHERFVEAVNQLGGADKATPKTVMKLMGIPGLTLYHLKSHLQqKYRL 106
79************************************************649**8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.137 | 48 | 108 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-29 | 49 | 107 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.33E-14 | 50 | 106 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.1E-21 | 51 | 107 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.9E-8 | 53 | 102 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 7.2E-11 | 151 | 173 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Pfam | PF14379 | 1.9E-7 | 214 | 235 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 429 aa Download sequence Send to blast |
MMIFKTGRVP SPNQATMFSS PRMSAPQERQ LFLQGGNGSR DSNLVLSTDA KPRLKWTHDL 60 HERFVEAVNQ LGGADKATPK TVMKLMGIPG LTLYHLKSHL QQKYRLSKNI HGQTTGGPNK 120 AVVMSAEKMP EPNPSNTSSI NITHQPNKNL HINEALQMQI EVQRRLHEQL EVHDPVYFQF 180 KQNGLSPKAD TTTAAQNLQR HHPMNSHDKD VGRLQLRIEA QGKYLQSVLE KAEETLGKQN 240 IGVVGLEAAK LQLSELVSTV SNQCLNLALA EMAEEQQGIT PKQQIQPHKL MGNCSIDSCL 300 TSCDGIQQRE PDMQNSELGL RPYNDSEFLK GNDDSENIKD HANLLFPTTT QQDTGKMMFP 360 VEGACNDLSM SVGIGGDTRD ELEKNAFQHD NLNYKLSSFS GDLDLNTQDE PDASTSIRHL 420 DLNGLSWT* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 5e-19 | 51 | 108 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 5e-19 | 51 | 108 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 5e-19 | 51 | 108 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 5e-19 | 51 | 108 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 5e-19 | 50 | 108 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
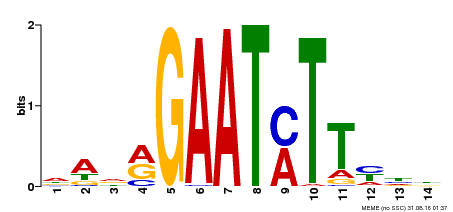
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010672720.1 | 0.0 | PREDICTED: myb-related protein 2-like isoform X1 | ||||
| Refseq | XP_010672724.1 | 0.0 | PREDICTED: myb-related protein 2-like isoform X1 | ||||
| TrEMBL | A0A0K9RSG6 | 0.0 | A0A0K9RSG6_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010672720.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 1e-102 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_000118-RA |



