 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_000139-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 377aa MW: 40588 Da PI: 8.8738 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 138.2 | 2.4e-43 | 75 | 150 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+CqvegC++dls+ak y++rhkvC++hsk+p v+v+gleqrfCqqCsrfh+l efD++krsCrrrLa+hnerrrk+
AHYPO_000139-RA 75 RCQVEGCKVDLSDAKPYYSRHKVCSMHSKSPLVIVNGLEQRFCQQCSRFHRLPEFDQGKRSCRRRLAGHNERRRKP 150
6*************************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.6E-34 | 71 | 137 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.565 | 73 | 150 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.22E-41 | 74 | 154 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 8.0E-33 | 76 | 149 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:2000025 | Biological Process | regulation of leaf formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 377 aa Download sequence Send to blast |
MDTTTSKYQT FVEGSSKTKP LSSSGLSDSL NGLKFGQKIY FEDVGGTSLG KSCSSSSISS 60 KKGGKGVLQS GLPPRCQVEG CKVDLSDAKP YYSRHKVCSM HSKSPLVIVN GLEQRFCQQC 120 SRFHRLPEFD QGKRSCRRRL AGHNERRRKP PPGSLLSSRL GRLSSSLFDD KTNRSGGFLL 180 DFTSYSRDSK TDLWLGYNTS EQVSGIHSRS NMWPGNSIDP SSKVFPQGSA SGNVFSFPSG 240 ECNTGVSSDS SCALSLLSNQ NWSSGNRASD TGLGNLMNID GVAVSQCPAA HTVTVAHLPS 300 SSSLWGFKGN NDPNCGLHAV PSGHPGLGQT SQPHFSGQFH GDLMMEQVPD GGRHYMDVEH 360 SRVYDSTTSN HVNWSI* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-30 | 70 | 149 | 6 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May be involved in panicle development. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00307 | DAP | Transfer from AT2G42200 | Download |
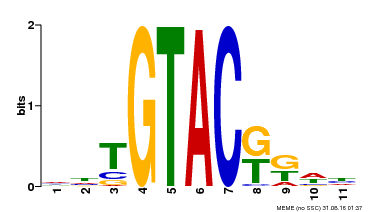
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Negatively regulated by microRNAs miR156b and miR156h. {ECO:0000305|PubMed:16861571}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021724004.1 | 1e-171 | squamosa promoter-binding-like protein 17 isoform X2 | ||||
| Swissprot | A3C057 | 1e-61 | SPL17_ORYSJ; Squamosa promoter-binding-like protein 17 | ||||
| TrEMBL | A0A0K9QU54 | 1e-158 | A0A0K9QU54_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010672689.1 | 1e-170 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G42200.1 | 3e-37 | squamosa promoter binding protein-like 9 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_000139-RA |



