 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_000259-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 405aa MW: 45842.7 Da PI: 6.6776 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 89.9 | 2.7e-28 | 71 | 156 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkr.tsessstcpyfdqle 86
W ++e++ Li++rrem++ ++++k++k+lWe++s+kmrergf rsp++C++kw+nl k++kk k++ +++ + + s +++y++++e
AHYPO_000259-RA 71 WVQDETQNLIALRREMDSLFNTSKSNKHLWEQISAKMRERGFDRSPTMCTDKWRNLLKEFKKAKQQLGNGcNGNGSAKMSYYKEIE 156
**********************************************************************7788899******998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50090 | 8.664 | 63 | 127 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-5 | 64 | 129 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 6.2E-6 | 67 | 129 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.22E-30 | 69 | 134 | No hit | No description |
| Pfam | PF13837 | 5.5E-20 | 70 | 158 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 405 aa Download sequence Send to blast |
MYISDTNHNP TTTTTTTTKP QSSSIEFSDG DHHNPHDMMI EVVTTHHHHD HHRLPTTGTT 60 SSGPKKRAET WVQDETQNLI ALRREMDSLF NTSKSNKHLW EQISAKMRER GFDRSPTMCT 120 DKWRNLLKEF KKAKQQLGNG CNGNGSAKMS YYKEIEEIIR ERNRCSSSSN VAGYRSPSHN 180 NGGVNHSSKD DPFLQFSDKG IDDATISFGP IEDNGRTALN LERSLDHEEH SLAITAADAV 240 AGGGVPPSFW RETPGNGDGT NSYGGRVITV KWGNYTRRIG IDGTAEAIKD VIRSTFGLRT 300 KRAFWLEDEY QVVRPLDRDM PLGTYALHLD EGVTIKLCHY EESKHISYPS PTISHTEEKT 360 FYTEDDFREF MHRCGWSCLR ELNGYRNIDN LDDLRPDALY RGIV* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 1e-43 | 65 | 154 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00640 | PBM | Transfer from MDP0000164819 | Download |
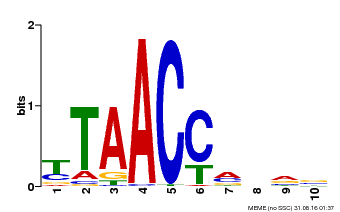
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_021746780.1 | 0.0 | trihelix transcription factor GT-4-like | ||||
| Swissprot | Q9FX53 | 1e-156 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A0K9RAW0 | 0.0 | A0A0K9RAW0_SPIOL; Uncharacterized protein | ||||
| STRING | XP_010682875.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 1e-143 | Trihelix family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_000259-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



