 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | AHYPO_001499-RA | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; Caryophyllales; Amaranthaceae; Amaranthus
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 932aa MW: 103533 Da PI: 6.5922 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 132.2 | 1.7e-41 | 144 | 221 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqve+C dls+ak+yhrrhkvCevhska+++lv++++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+q+
AHYPO_001499-RA 144 ICQVEDCGIDLSKAKDYHRRHKVCEVHSKASQALVANVMQRFCQQCSRFHVLQEFDEGKRSCRRRLAGHNKRRRKTQP 221
5**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 4.5E-33 | 137 | 206 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.017 | 142 | 219 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 2.35E-38 | 143 | 224 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 1.2E-29 | 145 | 218 | IPR004333 | Transcription factor, SBP-box |
| SMART | SM00248 | 290 | 704 | 734 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF48403 | 9.02E-8 | 707 | 814 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.7E-7 | 708 | 817 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.13E-8 | 709 | 810 | No hit | No description |
| SMART | SM00248 | 1.4 | 757 | 787 | IPR002110 | Ankyrin repeat |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 932 aa Download sequence Send to blast |
MEQAHRFHSI DPSELRVMGK RGLEWDLNDW KWDGDLFIAR PSNSIPTNFQ GQQFIPVLAG 60 NSSNCSSSCS DDVQYGVDDG RRELERKRRV IVVEEDGLGE SGSLTLKLGG NCNPAIDRER 120 EGVNWEGTSG KKAKVTGGSS NRAICQVEDC GIDLSKAKDY HRRHKVCEVH SKASQALVAN 180 VMQRFCQQCS RFHVLQEFDE GKRSCRRRLA GHNKRRRKTQ PETAVQGSSV NDEQTSSVLL 240 MSLLKILSDM HANNRPDQAT DQDLVVQLLR GLANPSLHMG KGLSGLQPES QKLLNGGIAN 300 DNGHSKKMSA FLSNDNRNPP RHIDQHVPLP DSEIPGKGVY AANNRGVDMQ AGSLFPIKDS 360 PPGYSTEGRM KLNNFDLNDV YVDSDDGIED LERSPVNGNF ATGSVDFPLW ARQDSHQSSP 420 PQTSGNSDSA SALSPSSSSG EASILDWLAH SPTDIESYIR PGCIILTIYL RLTESLWEEL 480 CSDLSSRLTR LFDITDDSFW RTGWVYVRVQ NQIAFVHNGQ VVLDTSMSLD NNSCSRILSV 540 MPIAVSMDER AHFEVRGFNL SHSTTSYVFD QKICRVEDLG LSSSFFPFIV AEKDVCSEIR 600 TLESVLEPKN SDQDVYEANK NIESWRQAMD FIHEMGWLLH RSHMKSRLAD LDPSTVVFSF 660 RRYKWLMDFS MDHDWCAVVK KLLDILVAGT VGSGEHPSLK VALSEMGLLH RAVRRNSRSM 720 VELLLRYVPA TLSEEFTSVA ESSQEKFLFR PDAQGPGGLT PLHVAAGRDG SDDVLDALID 780 DPGKIGIETW KNSRDSTGAT PEDYARLRGH YSYIHVVQRK MPRSSASGHV VLDIPGEPSV 840 AAPKHDGAVS FEIGRSASFA LNQNCKMCDH KKMLAYYGRS TKTSLAYRPL MLSMVGIAAV 900 CVCVALLFKS MPNVVCLFQP FRWELLNYGS S* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-32 | 136 | 218 | 2 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
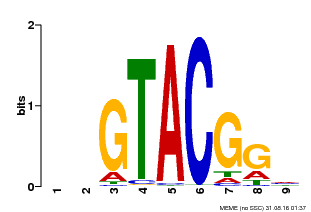
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00633 | PBM | Transfer from PK06791.1 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY095310 | 1e-87 | AY095310.1 Amaranthus palmeri x Amaranthus rudis clone 21B.206 hybrid progeny specific sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010678887.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | A0A0J8BDT7 | 0.0 | A0A0J8BDT7_BETVU; Uncharacterized protein | ||||
| STRING | XP_010678887.1 | 0.0 | (Beta vulgaris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | AHYPO_001499-RA |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



